Plant Immunity

- Professor
- SAIJO Yusuke

- Assistant Professor
- YASUDA Shigetaka

- INOUE Kanako

- Dominguez John Jewish

- Labs HP
- https://bsw3.naist.jp/saijo/
Outline of Research and Education
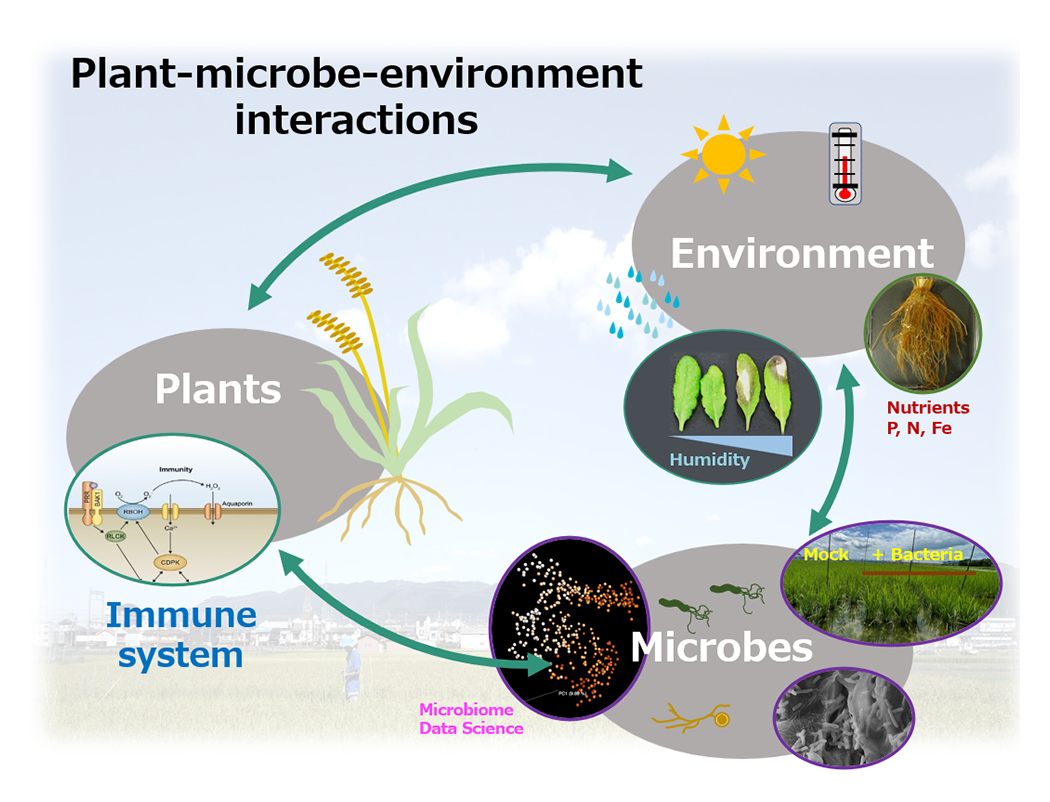
In nature, plants harbor a diverse community of microbes, ranging from mutualistic symbionts to pathogens. The dynamics and outcomes of plant-microbe interactions, including crop disease epidemics, are profoundly influenced by environmental factors, such as light, temperature, water, and nutrient availability. Our research aims to uncover the mechanisms by which plants perceive and integrate biotic and abiotic signals to regulate their microbial associations in fluctuating environments. Our key research areas include: (1) immune receptor signaling and the crosstalk between biotic and abiotic stress responses, (2) functional significance and infection strategies of mutualistic and pathogenic microbes, and (3) understanding and harnessing plant-associated mutualistic microbes. Through these studies, we seek to reveal fundamental principles of host-microbe interactions and contribute to the advancement of sustainable agriculture.
Major Research Topics
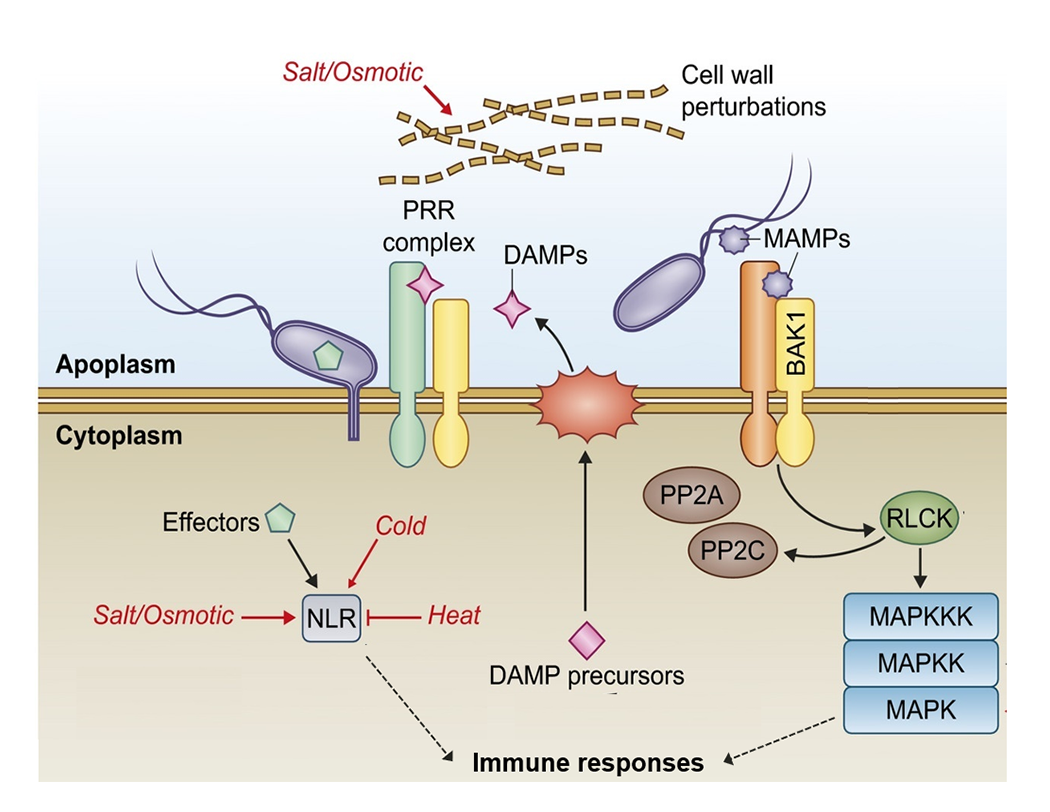
- Danger sensing and signaling in plant immunity
- Signal integration between biotic and abiotic stress responses in plants
- Infection strategies and regulatory mechanisms of mutualistic and pathogenic microbes in plants
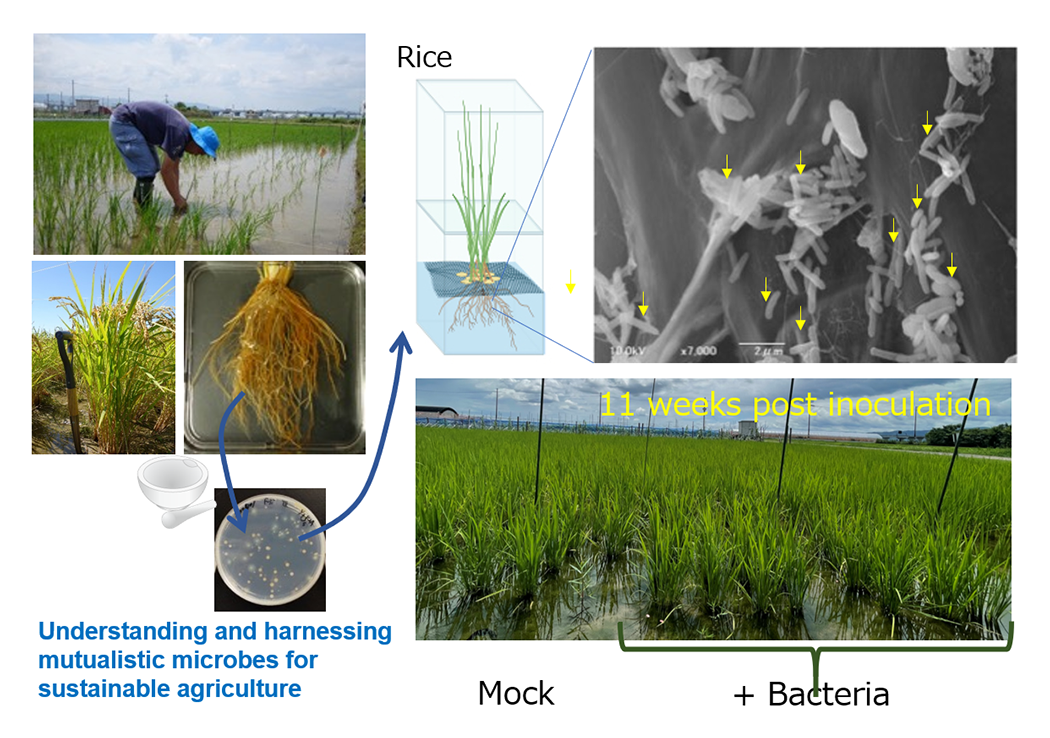
- Harnessing plant-associated mutualistic microbes for sustainable agriculture
References
- Yasuda et al, bioRxiv 2025 doi: 10.1101/2025.01.04.63131
- Adachi et al, bioRxiv 2024 doi: 10.1101/2024.09.02.610732
- Tanaka et al, Mol Plant Microbe Interact 38, 2025 in press
- Okada et al, Plant J 120, 2639-2655, 2024
- Inoue et al, J Plant Res 137, 343-357, 2024
- Hiruma et al, Nature Commun 14, 5288, 2023 doi: 10.1038/s41467-023-40867-w.
- Okada et al, New Phytologist, 229, 2844-2858, 2021
- Saijo & Loo, New Phytologist, 225, 87-104, 2020 Tansley Review
- Saijo et al., Plant J., 93, 592-613, 2018
- Shinya et al., Plant J., 94, 4, 626-637, 2018
- Yasuda et al, Curr Opin Plant Biol, 38, 10-18, 2017
- Ariga et al, Nature Plants, 3, 17072, 2017
- Yamada et al, Science, 354, 1427-1430, 2016
- Espinas et al, Front. Plant Sci., 7, 1201, 2016
- Yamada et al, EMBO J., 35, 46-61, 2016
- Ross et al., EMBO J., 33, 62-75, 2014
- Tintor et al., Proc Natl Acad Sci U S A, 110, 6211-6216, 2013

 NAIST Edge BIO
NAIST Edge BIO