Microbial Molecular Genetics


Outline of Research and Education
At our research group, we have been studying (1) how genomic instability is caused when an antimicrobial reagent induces replication inhibition and cell death, and (2) how microbial biofilms that hamper drug treatments of microbial infection are formed and disassembled. We also put strong emphasis on the international education of young students who are highly interested in these research themes.
Major Research Topics
Genomic instability under low levels of nucleotides (Fig. 1−3)
Replication fork movement on chromosomal DNA is impeded by numerous natural obstacles including spontaneous DNA lesions, unusual DNA structures, DNA-binding proteins, RNA polymerases and depletion of nucleotide pools. Inhibition of replication fork progression (replication stress) potentially cause genomic instability, characteristics of most cancers(Fig.1).
It has not been well elucidated where and how fork movement is blocked on the genome, and how the replication blockage leads to genomic instability. To approach these questions, it is important to understand molecular dynamics of replication fork movement on chromosome. As described in biology textbooks, basic molecular mechanisms of replication enzymes are well known. However, an overall picture of replication fork dynamics on the genomic DNA has not been fully understood yet in any organisms. We have recently succeeded in determining distribution of replication forks on the E. coli genome (Fig.2)
When cells are starved for the DNA precursor dTTP, they quickly lose viability, a phenomenon named thymine-less death (TLD). Although TLD has been exploited by popular anti-cancer and bactericidal drugs (5-fluorouracil and trimethoprim), the precise mechanisms of genomic instability and cell lethality in TLD have been unknown since its discovery more than 60 years ago. We have recently found that the E. coli genome is programmed to arrest replication fork at discreate regions near the replication origin under thymidine shortage and to produce genetic instability under thymidine starvation (Fig.3).
Biofilm formation by Bacillus subtilis(Fig. 4)
In environment, most bacteria are found in surface-associated multicellular communities, termed as biofilms. In biofilms, bacterial cells are encased in a self-produced matrix of extracellular polymeric substances, which protects resident cells from various antimicrobials and stresses. Biofilms are therefore notoriously difficult to eradicate and sometimes pose a serious threat to public health and our environment. However, molecular mechanisms for biofilm formation remain to be elucidated. To address the issues, we are exploring mechanisms of biofilm formation by the Gram-positive bacterium Bacillus subtilis. We are also interested in biofilm-related phenomena such as bacterial competition and antibiotic production.
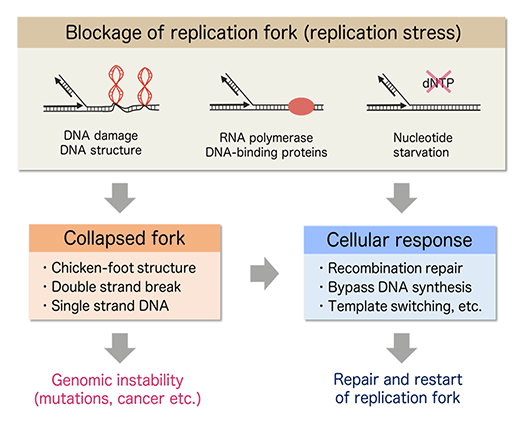
Because blocked fork results in DNA damage leading to genomic instability, cells deal with the inhibition by various repair mechanisms.
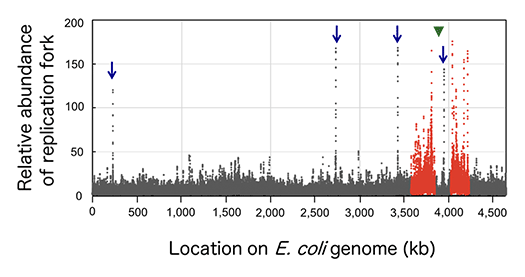
Slow replication occurs at rrn genes (blue arrows) and the regions (red dots) proximal to the replication origin (a green triangle).
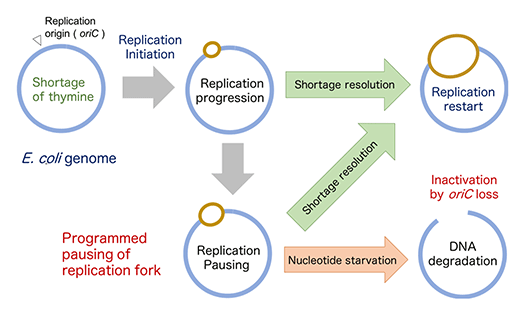
The E. coli genome is programmed to pause replication and induce genome instability near the replication origin under nucleotide shortage.
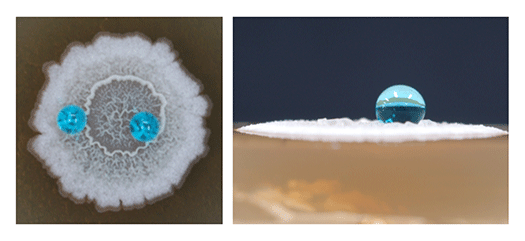
(Left) B. subtilis forms architecturally complex colony biofilms. (Right) The biofilms exhibit surface water-repellency. Light blue circles are water drops.
References
- T.M. Pham et al., Mol. Microbiology, 90, 584-596, 2013
- K.W. Tan et al., Nucleic Acid Res., 43, 1714-1725, 2015
- M.T. Akiyama et al., Genes to Cells, 21, 907-914, 2016
- K. Kobayashi, J. Bacteriology, 201, e00712-18, 2019
- K. Kobayashi and Y. Ikemoto, PLoS Genetics, 15, e1008232, 2019
- T. Kimura and K. Kobayashi, J. Bacteriology, 202, e00120-20, 2020

