Plant Metabolic Regulation

- Professor
- DEMURA Taku

- Assistant Professor
- KUNIEDA Tadashi

- MIZUTANI Miya

- Labs HP
- https://bsw3.naist.jp/demura/
Outline of Research and Education
Our laboratory engages in research and education pertaining to the biotechnology needed to resolve the issues facing human beings in the 21st century, such as food, environment, and energy. Especially we are exploring the mechanisms of gene expression regulation for woody cell differentiation using omics technology to develop novel biotechnological tools for the establishment of a sustainable society.
Major Research Topics
Molecular mechanisms governing xylem cell differentiation
Xylem functions in conduction of water and minerals throughout the plants, and supports the plant body. One of the features of xylem cells is development of secondary wall structure between plasma membrane and (primary) cell wall. Since woody biomass derived from xylem cells, xylem vessels and fiber cells, is one of important resources of land plant biomass, modifications of molecular mechanisms for xylem cell differentiation should be important strategies to improve plant biomass resources.
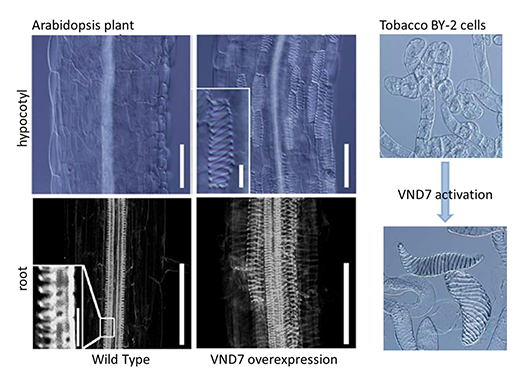
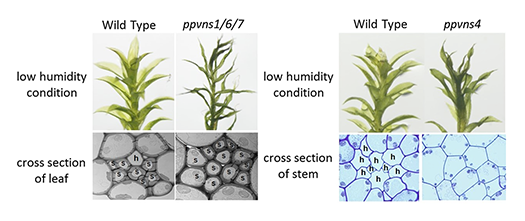
We identified a key regulator of the xylem vessel differentiation, Arabidopsis VND7 (VASCULAR-RELATED NAC-DOMAIN7), which is a plant-specific NAC domain transcription factor (Fig. 1). To understand the molecular mechanism by which xylem vessel formation is regulated, we have been characterizing VND7 and its homologs through various approaches. Recently we investigated the VND-homologous genes of moss Physcomitrella patens, and found that the VND-homologous genes function in the differentiation of water-conducting and supporting cells in the moss (Fig. 2). The VND-based molecular system is thus conserved among current land plants widely, suggesting that our findings can be applied to wide-range land plants to modify woody biomass.
We are also conducting genomics, transcriptome, proteome and metabolome studies to reveal the molecular system of plant biomass biosynthesis, using not only model plants but also non-model practical plants. Based on the obtained information, the designed transgenic poplars were generated and tested for the useful trains, such as higher stress tolerance, plant growth, and woody biomass accumulation (Fig. 3). These studies will give us the insights into effective biotechnological strategy to improve the quality and quantity of woody biomass.
Mechanism of plant mechanical optimization system
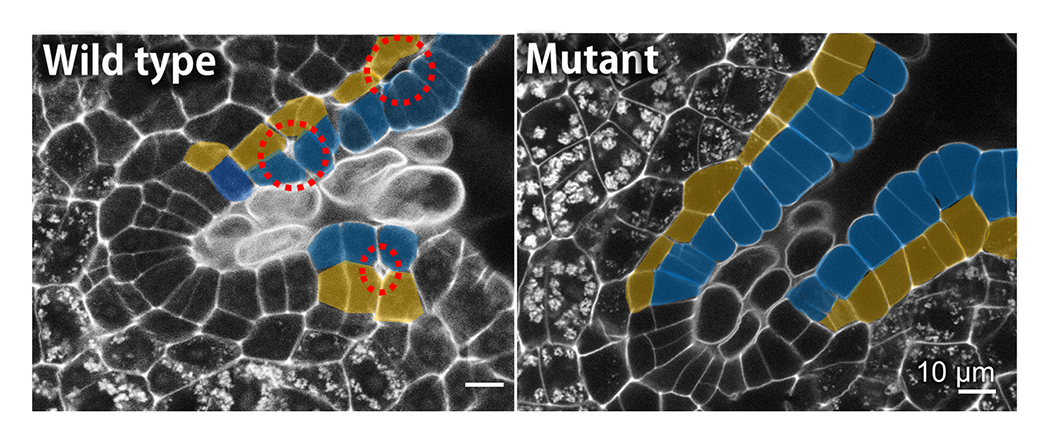
Plants change their structures into mechanically optimal shapes in the process of development and environmental response. We are analyzing the mechanical optimization system of plants on various scales (molecule-cell-tissue-organ) and elucidating its mechanism (Fig. 4). We are performing the mechanical analysis using atomic force microscope (AFM), the 3D/4D structural analysis using time-lapse camera, micro X-ray CT and IoT devices (Raspberry Pi, etc.), and these analyses combined with the mathematical analysis. Based on the research findings, we are aiming to develop the application for establishing next-generation bio-basic technologies such as sophistication of plant function, and for creating sustainable architecture that is in harmony with various environmental factors such as earthquakes, typhoons, and temperature.
Molecular mechanism of the endomembrane trafficking underlying plant cell wall formation.
Cell wall polymers are secreted to the outside of the plasma membrane via endomembrane organelles after their biosynthesis. In order to understand cell wall formation, we are studying the factors that are involved in the endomembrane system for secretion of plant cell wall polysaccharides, focusing on their spatiotemporal distribution inside cells (Fig. 5). Our findings can contribute to establish technologies that efficiently control intracellular transport of substances by the endomembrane trafficking system.

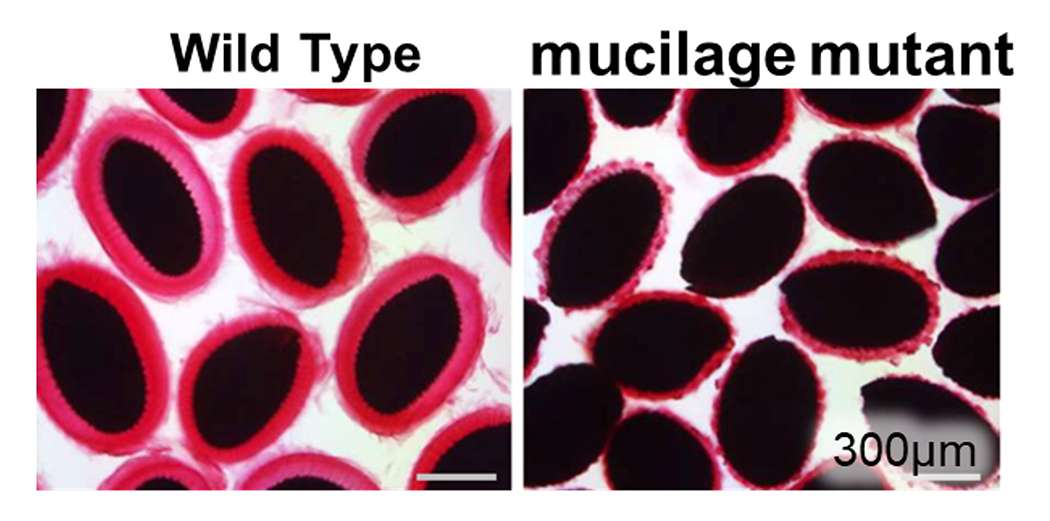
The mucilage is one of model cell walls for studying cell wall formation. We have identified several factors that are required for mucilage formation. Red and black regions in the pictures corresponds mucilage and seeds, respectively.
References
- Takahara et al., Plant Physiol., kiad105, 2023
- Nakata M.T. and Takahara M., Int. J. Mol. Sci., 23, pp10240, 2022
- Nakano Y. et al., Front. Plant Sci., 13, 819360, 2022
- Hirai R. et al., Front. Plant Sci., 13, 825810, 2022
- Akiyoshi et al., Plant Cell Physiol., 62, 1963–1974, 2021
- Terada et al., Plant Mol. Biol., 106, 309-317, 2021
- Nakata et al., Front. Plant Sci., 12, pp654655, 2021
- Kunieda T. et al., Plant Cell Physiol., 61, 308-317, 2020
- Tsugawa S., J. Theor. Biol., 486, 110092, 2020
- Yoneda A. et al., Plants, 9, 604, 2020
- Akiyoshi N. et al., Tree Biol., 40, 704–716, 2019
- Hirai R. et al., Plants, 9, 39, 2019
- Tamura T. et al., Plant J., 100, 298-313, 2019
- Ohtani M. et al., Curr. Opin. Biotech, 56, 82-87, 2019
- Chiam NC. et al., Plant Cell Physiology, 60, 2000 -2014, 2019
- Kubo M. et al., Nucleic Acids Res., 47, 4539-4553, 2019
- Saelim L. et al., J. Plant Res., 132, 117 -129, 2019
- Takenaka Y. et al., Plant Cell, 30, 2663-2676, 2018
- Ohtani M. Front. Plant Sci., 8, 2184, 2018
- Tan T. et al., Plant Physiol, 176, 773-789, 2018
- Noguchi M. et al., Plant Biotechnol, 35, 31-37, 2018
- Ohtani M. et al., Plant Signal Behav, 13, e1428512, 2018
- Kawabe H. et al., Plant Cell Physiol, 59, 17-29, 2018
- Yu X. et al., J Plant Res, 130, 929-940, 2017
- Ohtani M., J Plant Res, 130, 57-66, 2017
- Yu X. et al., Mol Breed, 37, 57, 2017
- Ohtani M. et al., J Exp Bot, 68, 17-26, 2017
- Ohtani M. et al., Plant Physiol, 172, 1612-1624, 2016
- Watanabe Y. et al., Science, 350, 198-203, 2015
- Rejab NA. et al., Plant Biotechnol, 32, 343-347, 2015
- Endo H. et al., Plant Cell Physiol, 56, 242-54, 2015
- Ohtani M. et al., J Plant Res, 128, 28, 371-80, 2015
- Nakano Y. et al., Front Plant Sci, 6, 288, 2015
- Ohtani M., J Plant Res, 128, 361-369, 2015
- Xu B. et al., Science, 343, 1505-1508, 2014
- Matsuura H. et al., Plant Cell Physiol., 54, 474-483, 2013
- Ohtani M. et al., Plant Cell, 25, 2056-2069, 2013
- Matsui T. et al., Transgenic Res., 20, 735-748, 2011
- Ohtani M. et al., Plant J. 67, 499-512, 2011
- Yamaguchi M. et al., Plant J, 66, 579-590, 2011

 NAIST Edge BIO
NAIST Edge BIO