Structural Life Science

- Professor
- TSUKAZAKI Tomoya

- Assistant Professor
- MIYAZAKI Ryoji

- KITANO Ken

- KOHGA hidetaka

- Labs HP
- https://bsw3.naist.jp/tsukazaki/en.html
Outline of Research and Education
Biological phenomena are driven by a variety of macromolecules, including proteins, RNA, and DNA. Our research aims to elucidate the molecular mechanisms underlying their dynamic structural changes at the atomic level. To achieve this, we integrate innovative research methods with structural biology approaches, conducting fundamental studies to deepen our understanding of these processes.
Our laboratory provides an environment where researchers can fully immerse themselves in their work, fostering high-quality research output. Conducting research requires diverse skills, from experimental design to presenting findings. We encourage graduate students to develop these skills through their research activities, helping them grow as researchers and as valuable members of society.
Major Research Topics
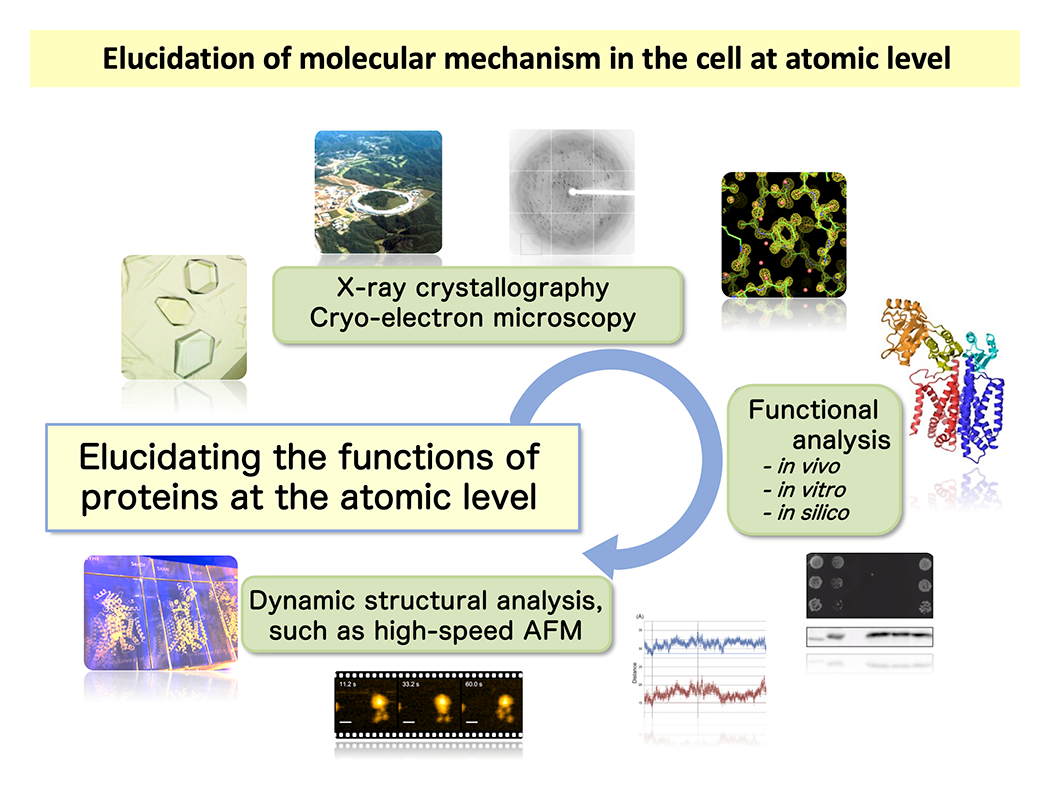
Figure 1 illustrates our research workflow. We begin by determining the atomic and molecular level structures of proteins. Once we obtain detailed structural information, our perspective broadens significantly, allowing us to gain valuable insights into how the target protein functions. This is the most significant advantage of elucidating structural details. Next, we formulate working hypotheses based on the structural data and validate them through functional analyses. In recent years, we have also incorporated single-molecule and in vivo dynamics analyses to visualize proteins in action within living systems. Our research aims to uncover the fundamental principles of molecular function with the potential to shape the future of biomedical science. We study a variety of proteins, including drug targets, with a particular focus on transport proteins and motor proteins, which are essential for cellular processes and disease mechanisms.
Visualization of the Operational Mechanism of the Sec Membrane Protein Complex at the Atomic Level
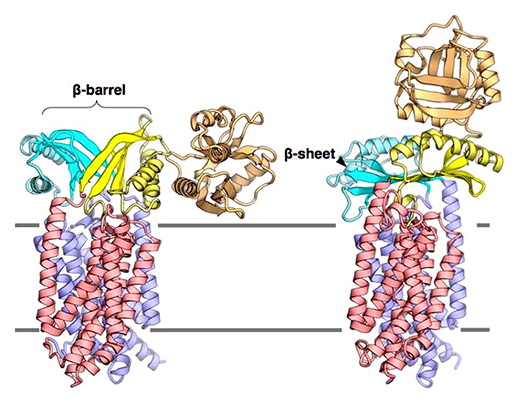
The Sec membrane protein complex is an essential machinery found in the cytoplasmic membrane of bacteria and the endoplasmic reticulum membrane of eukaryotic cells, facilitating the translocation of newly synthesized proteins across membranes. The process of protein translocation has been extensively studied, with key milestones including the "signal hypothesis" proposed by Blobel et al. in 1975, which later led to a Nobel Prize. Our research focuses on elucidating the detailed structures of individual Sec factors and conducting functional analyses based on structural information. Through these efforts, we have uncovered the structural changes that occur during protein translocation. We aim to expand our research by resolving the full atomic-level structure of the Sec membrane protein complex and tracking its time-dependent reactions to unravel its dynamic operational mechanism in greater detail.
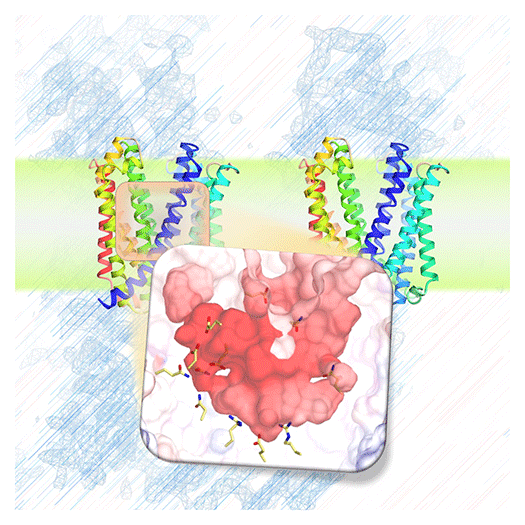
Unraveling the Intricate Mechanisms of Ion Transporters
Cells are compartmentalized by biological membranes, which separate the external environment from the intracellular space and organelles. Through the activity of membrane proteins, cells regulate the uptake and efflux of substances such as drugs, facilitate signal transduction, and drive energy production. To fully understand the molecular mechanisms of these membrane proteins, it is essential to determine their three-dimensional structures. These structural insights provide fundamental knowledge for deciphering biological phenomena and advancing biomedical research.
Cutting-Edge Analysis Using Advanced Technologies
To investigate protein movements in real time, we employ cutting-edge analytical techniques. These include the PiXie (pulse-chase and in vivo photo-cross-linking experiment) method for in vivo protein dynamics analysis and collaborative studies utilizing high-speed atomic force microscopy (AFM) to observe the dynamics of individual protein units. Additionally, we use high-performance cryo-electron microscopy (cryo-EM) to capture structural snapshots at near-atomic resolution. By integrating these state-of-the-art technologies, we aim to gain a deeper understanding of protein function and dynamic behavior, advancing our knowledge of fundamental biological processes.
References
- Kohga et al., Sci. Adv., 11, eady8083, 2025
- Miyazaki et al., Cell Rep., 44, 116013 2025
- Kanaoka et al., Nat. Commun., 16, 225, 2025
- Ikei et al., PLoS Biol., 22, e3002601, 2024
- Tanaka et al., Sci. Adv., 6, eaba7637, 2020
- Furukawa et al., Cell Rep., 19, 895-901, 2017
- Tanaka et al., Cell Rep., 13, 1561-1568, 2015
- Kumazaki K. et al., Nature, 509, 516-520, 2014
- Tsukazaki T. et al., Nature, 474, 235-238, 2011
- Tsukazaki T. et al., Nature, 455, 988-911, 2008

 NAIST Edge BIO
NAIST Edge BIO