Research

Elucidation of protein dynamics at atomic and molecular levels
There are a number of proteins involved in life phenomena.
By combining structural biological analysis with new research techniques, we aim to advance our understanding of molecular mechanisms caused by dynamic protein structural changes.
Our current research focuses mainly on the transport of proteins and small molecules.
Main Research Interests
X-ray crystallography and single-particle cryo-electron microscopy have enabled one after another the determination of the structures of biomolecules.
In order to precisely understand fundamental biological phenomena, we are actively incorporating the latest research techniques, including dynamic structure analysis, into our fundamental research based on structural biology.
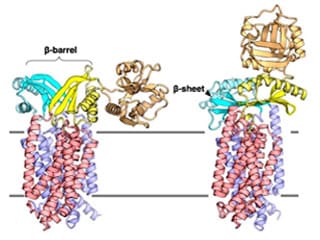
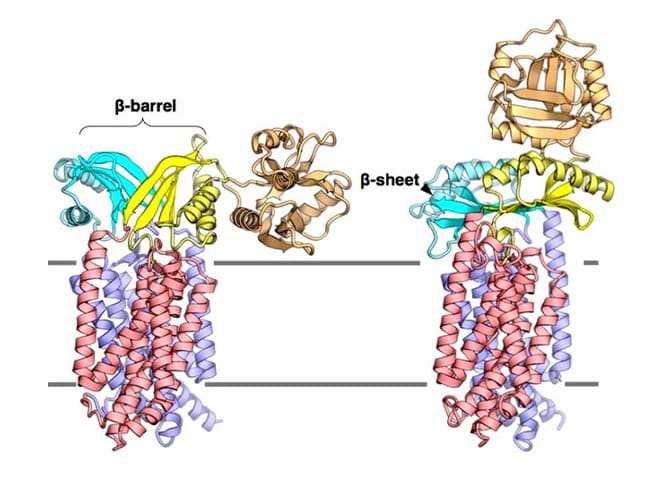
Ecluciation of Mechanism of Sec Membrane Protein Complex
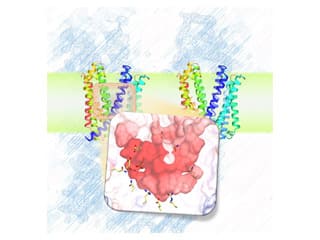
The figure shows the two conformational structures of SecDF, a motor protein that drives protein transport. These dramatic conformational changes and functional relationships are the subjects of our studies.
Elucidation of Precise Mechanisms of Transporters and Channels
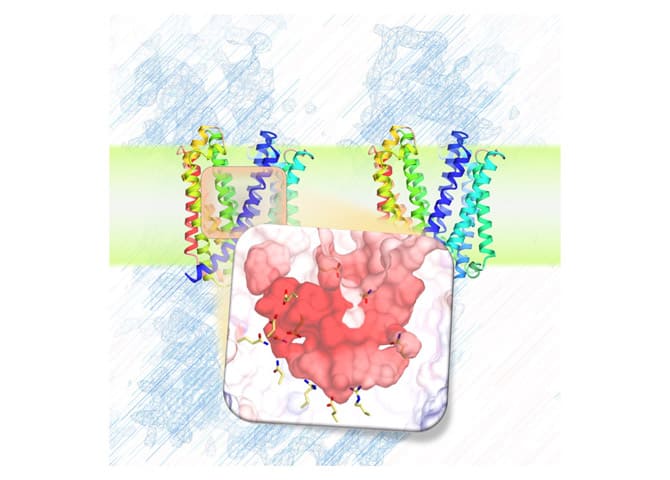
Across the membranes, transporters can transport drugs and information, generate energy, etc. We are studying their molecular mechanisms in detail.
Analysis Using the Latest Technology
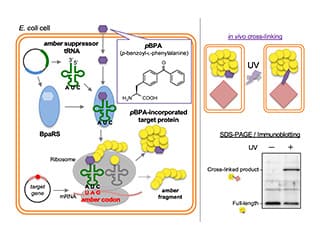
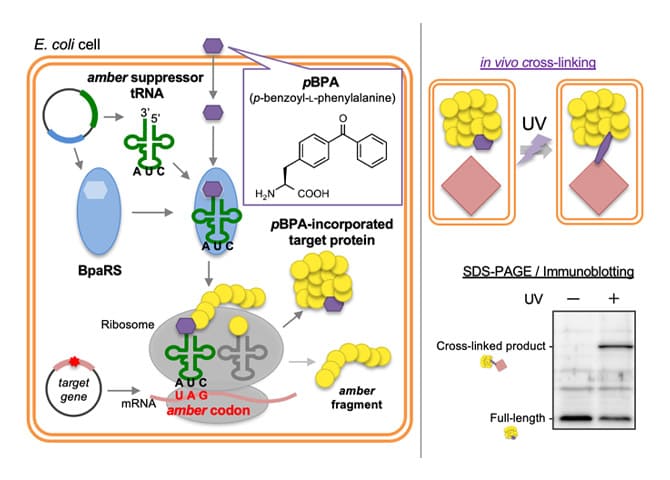
We are actively conducting research using the latest technologies, such as in vivo protein dynamics analysis using the PiXie method (see figure), single unit dynamics observation using high-speed atomic force microscopy, and high-performance cryo-electron microscopy.
What's New
- Phage lysis protein LysM acts as a wedge to block MurJ conformational changes.
- MurJ/LysM complex structure
Kohga et al., Sci. Adv. (2025)
- Structural basis of a GatC ortholog transporter in the bacterial phosphotransferase system.
- X-ray and cryo-EM structures of GatC
Takahashi et al., FEBS Letters (2025)
- Structural basis of lipopolysaccharide translocon assembly mediated by the small lipoprotein LptM.
- LptDEM structure and LptM function.
Miyazaki et al., Cell Reports (2025)
- We captured the process of a substrate protein through a membrane using HS-AFM.
-
A string-like protein (peptide) is being translocated by a single SecYEG-SecA.
Kanaoka et al., Nature communications (2025)
- The structure and function of YeeE-YeeD complex involved in thiosulfate ion transport.
-
We found the functional cooperation between YeeE and YeeD, which selectively transport thiosulfate ions in bacteria, and the enzymatic activity of YeeD. Bacteria can take up inorganic sulfur and convert it into organic compounds, contributing to the sulfur cycle on the Earth.
Ikei et al. PLoS Biol. 22, e3002601 (2024)
- Analyzing protein intermediate interactions in living E. coli cells using site-specific crosslinking
-
BepA directly interacts with the intermediates of the LptD in the vicinity of the β-barrel assembly machinery (BAM) complex to promote the maturation of a normal LptD or degrade an abnormal LptD. In this protocol, we present a method that combines chemical crosslinking and in vivo photo-crosslinking experiments to capture the interaction between BepA and LptD intermediates assembled into the outer membrane by the BAM complex.
Miyazaki and Akiyama. STAR Protocols 4, 102178 (2023)
- A multipoint guidance mechanism for β-barrel folding on the SAM complex.
- [Takeda et al., NSMB (2023)]
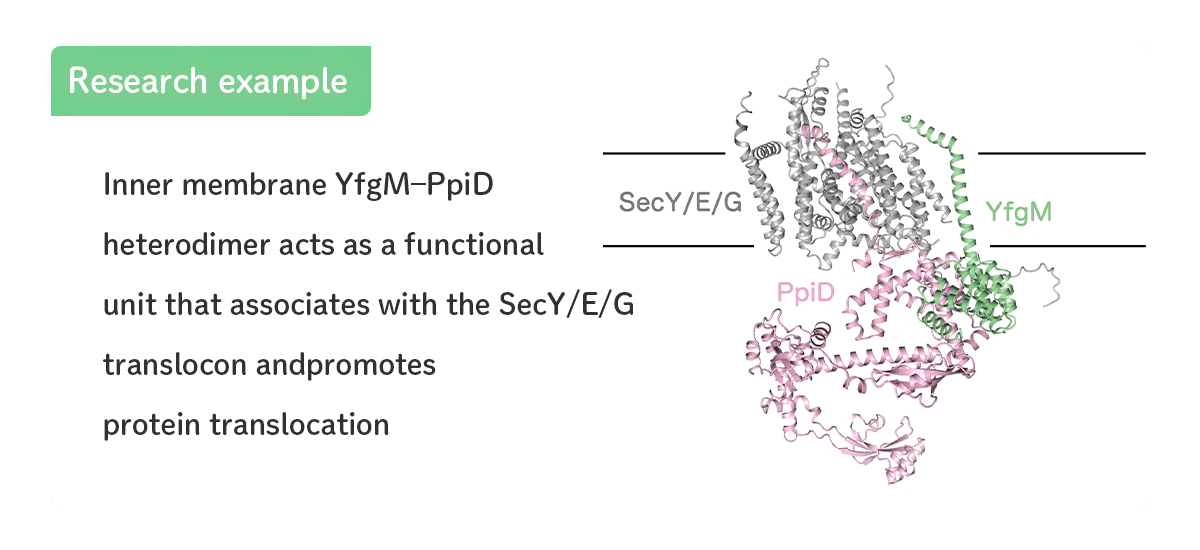
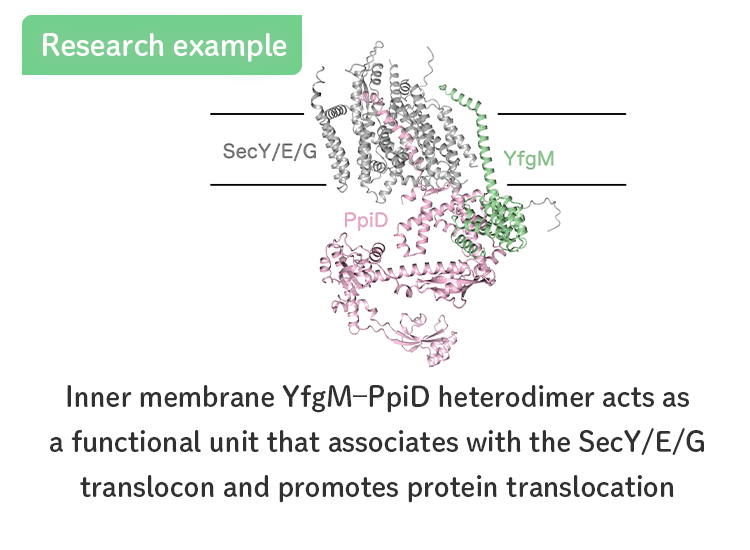
- Functional and structural analyses of YfgM, a novel factor involved in the protein translocation.
- [Miyazaki et al., JBC (2022)]
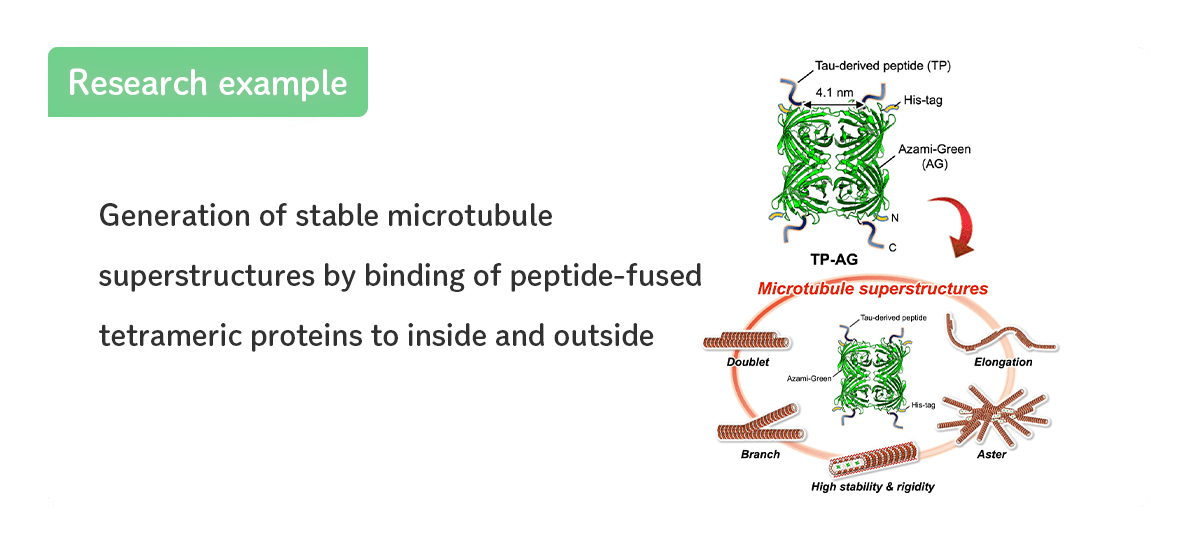
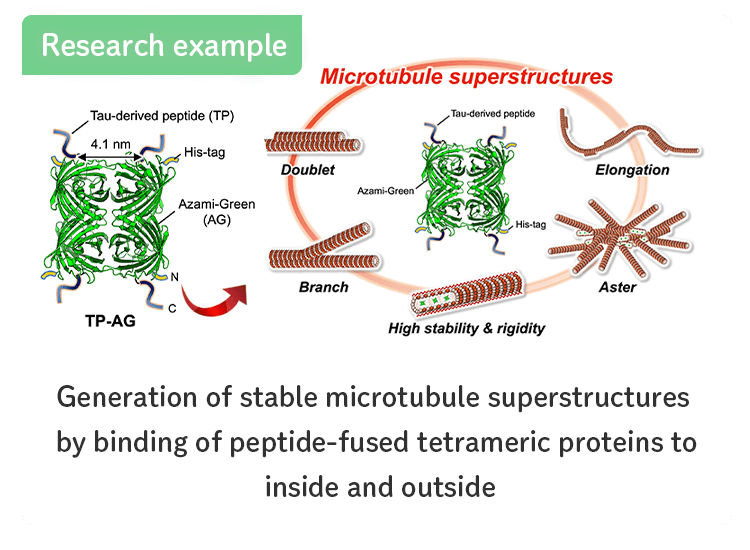
- Generation of stable microtubule superstructures...
-
〜分子ロボットなどのナノ材料への応用や繊毛・鞭毛の形成原理の解明に期待〜
バイオサイエンス領域[研究成果の紹介]はこちら
SCIENCE ADVANCES[Inaba et al., Sci. Adv.(2022)]
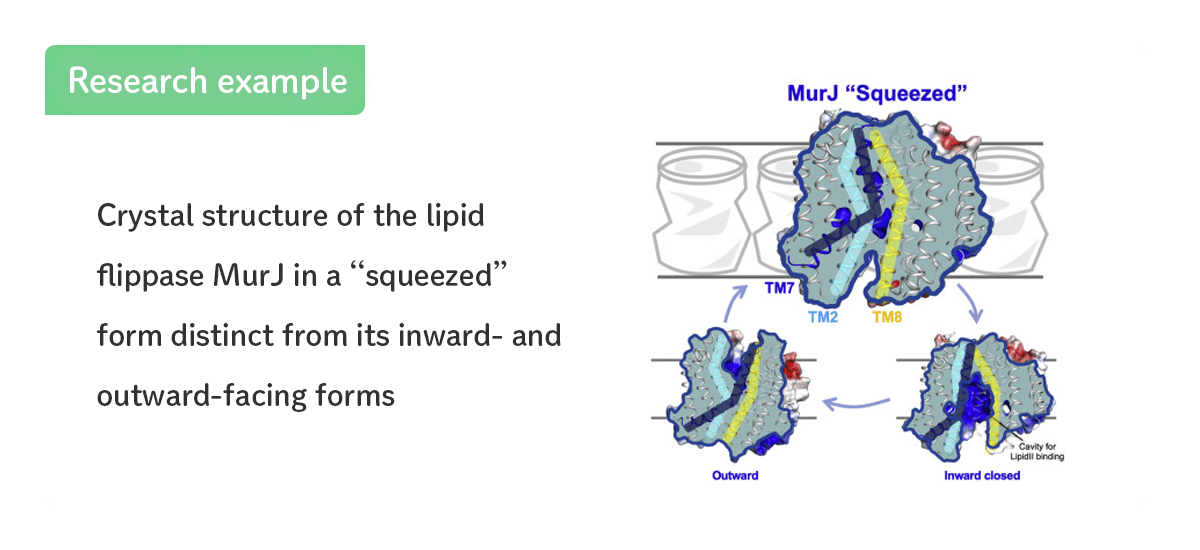
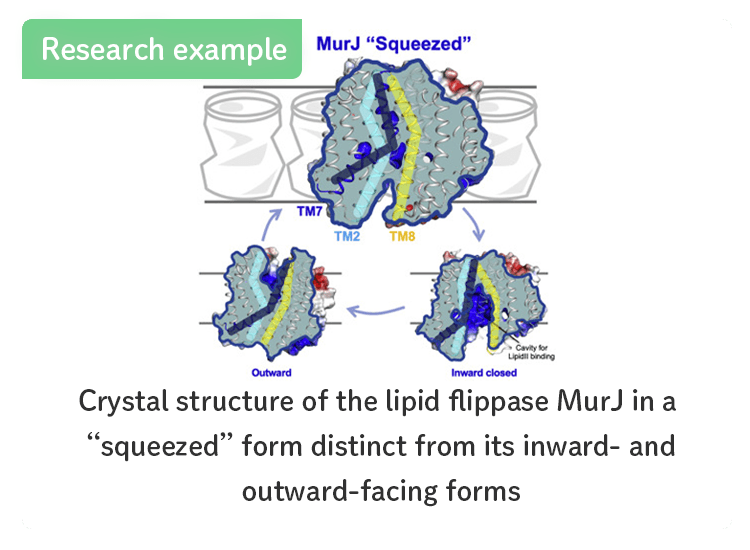
- Lipid flippase MurJ in a “squeezed” form.
- [Kohga et al., Structure (2022)]
- Crystal structures of a nicotine MATE transporter.
- [Tanaka et al., FEBS Letters (2021)]
- Characterization of nanoparticles by cryo-electron tomography.
- [Sheibani et al., Nat. Commun. (2021)]
- Reversible autoinhibitory regulation of BepA.
- [Daimon et al., PNAS (2020)]
- Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
- [Tanaka et al., Sci. Adv. (2020)]
- We investigated the architecture of the central pair complex of cilia by mass spectrometry.
- [Dai, Ichikawa et al., BPPB (2020)]
- The architecture of the Inner Junction region of doublet microtubule was revealed.
- [Khalifa, Ichikawa et al., eLife (2020)]
- YidC accelerates MPIase activity,
- [Sasaki et al., JBC (2019)]
- A new assist. prof., Hidetaka Kohga, joined us.
- Click here for details
- A new assist. prof., Ryoji Miyazaki, joined us.
- Click here for details