Molecular Medicine and Cell Biology

- Professor
- SUETSUGU Shiro

- Associate Professor
- NISHIMURA Tamako

- Assistant Professor
- KAWANA Hiroki

- Labs HP
- https://bsw3.naist.jp/suetsugu/
Outline of Research and Education
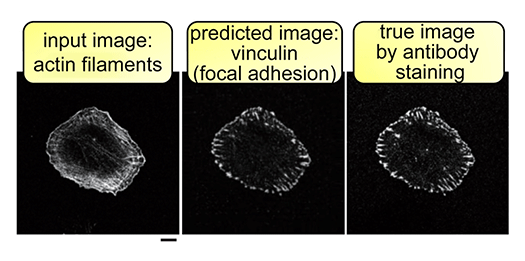
The cell membrane separates the inside and outside of the cell and is an indispensable structure for the cell to become a life. The cell membrane appears to be important for receiving the various stimuli that the cell receives. However, much remains unclear as to what combination of cell membranes and their binding proteins can shape cells and enable them to respond to stimuli. Our laboratory studies membrane-binding proteins that connect cell membranes to intracellular signaling in proliferation and morphological changes. The properties of the lipid molecules that make up the cell membrane are also investigated using membrane-binding proteins. Furthermore, using images showing the morphology of cells and images showing the localization of the above proteins, analysis of cell behavior, which is a change in cell morphology, is performed by deep learning.
Major Research Topics
Intracellular signaling depending on the morphology of the cell membrane and the proteins that form the morphology of the cell membrane, especially the association with cancer of the cell
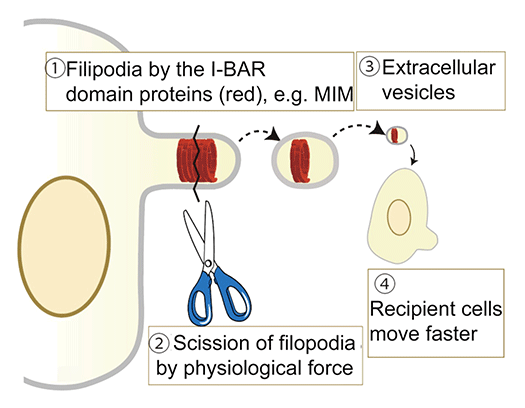
Cell morphological changes accompany important diseases such as cancer formation and various diseases. The same applies to cell differentiation and reprogramming. However, it is still unclear how the binding proteins of the lipid membrane are altered or regulated in the formation of organelles such as clathrin-coated pores, caveolae, filopodia, lamellipodia, and podosomes. We will clarify the role of the actin cytoskeleton and the membrane sculpting proteins, including the BAR domain-containing proteins, as well as lipid membranes in cell structure construction and intracellular signaling. In vitro reconstitution of membrane morphogenesis by membrane-binding proteins will be performed, and then the correlation between the reconstitution and the cell function will be investigated. Through such research, for example, we discovered the formation mechanism of extracellular vesicles that mediates intracellular communications.
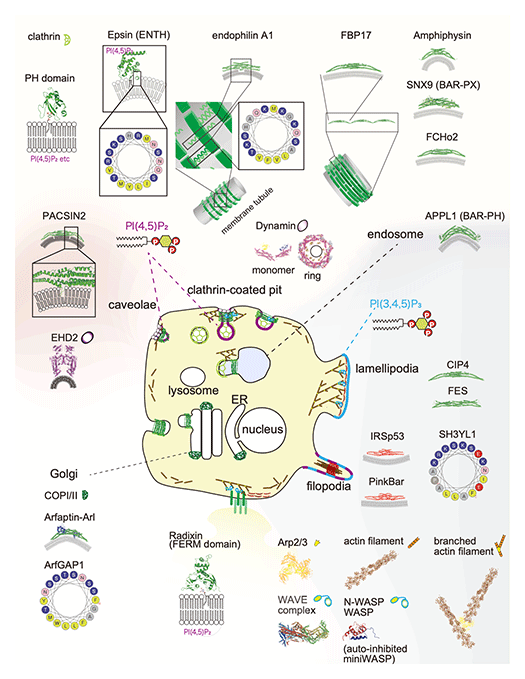
The BAR domain acts as a polymer of protrusions (including filamentous and lamellipodia) and submicron scale invagination (eg, clathrin-coated holes and caveolae) to form microstructures. Typical sizes for clathrin-coated pores and caveolae are 100-200 nm in diameter and 50-100 nm in diameter, respectively. The BAR domain can be approximated as a 20-25 nm arc with a diameter of 3-6 nm. The thickness of the membrane is approximately 5 nm.
References
- Hooi Ting Hu, et al., Front. Cell Dev. Biol., 12, 1422227, 2024
- Sim PF., et al., J Biochem, 175, 57-67, 2023
- Wan Mohamad Noor, WNI., et al., Sci Adv., 9, eadf5143, 2023
- Mukherjee, A. et al., Advanced Science (Weinh), 10, e2207368, 2023
- D'Angelo, G. et al., Nature Reviews Molecular Cell Biology, 24, 81-82, 2023
- Nishimura, T. el al., PLoS One., 17, e0271003, 2022
- Osuga, M. et al., Molecular Biology of the Cell, mbcE21010044, 2021
- Shigene, K., et al., Frontiers in Cell and Developmental Biology, 9, 635231, 2021
- Hu, HT., et al., STAR Protocols, 2, 100625, 2021
- Snider, CE. et al., Trends in Cell Biology, 31, 644-655, 2021
- Nishimura, T. et al., Developmental Cell, 56, 842-859, 2021
- Gusmira, A. et al, Journal of Cell Science, 133, jcs246785, 2020
- Kitamata, M. et al., Genes to Cells, 25, 187-196, 2020
- Hanawa-Suetsugu, K., et al., Nature Communications, 10, 4763, 2019
- Kitamata, M. et al., iScience, 17, 101-118, 2019
- Tachikawa, M. et al., Scientific Reports, 7, 7794, 2017
- Senju, Y., et al., Journal of Cell Science, 125, 2766-2780, 2015
- Takahashi, N. et al., Nature Communications, 5, 4994, 2014
- Suetsugu, S. et al., Physiological Reviews, 94, 1219-1248, 2014
- Suetsugu, S. and Gautreau, A., Trends in Cell Biology, 22, 141-150, 2012
- Senju, Y., et al., Journal of Cell Science, 124, 2032-2040, 2011
- Takano, K., et al., EMBO journal, 27, 2817-2828, 2008
- Shimada, A., et al., Cell, 129, 761-772, 2007

 NAIST Edge BIO
NAIST Edge BIO