Plant Stem Cell Regulation and Floral Patterning
- Professor
- ITO Toshiro

- Associate Professor
- YAMAGUCHI Nobutoshi

- Assistant Professor
- WADA Yuko

- Feng Yihong

- Labs HP
- https://bsw3.naist.jp/ito/
Outline of Research and Education
We are interested in the holistic view of gene regulation in plant reproduction, which leads to developmental robustness and coordination. We explore signaling and epigenetic control in stem cell maintenance, environmental response and fertilization. To reveal the molecular mechanisms, we use Arabidopsis as a model plant for genetic, reverse-genetic, biochemical and genomics approaches, as well as Brassicas and rice to study the conservation and diversification. Our students work at the frontiers of plant molecular genetics, developing their research, presentation and writing skills.
Major Research Topics
Proliferation, differentiation and senescence of floral stem cells
Flowers originate from self-renewing pluripotent stem cells in the floral meristems (Fig.1). In flower development, the stem cell activity is terminated in multistep pathways mediated by multiple transcription factors. The proliferation, differentiation and senescence of stem cells are regulated by a well-coordinated interplay of phytohormone signaling and epigenetic regulation, leading to spatiotemporal-specific gene regulation. We study downstream cascades of the key transcription factors controlling stem cell termination, flower organogenesis and senescence (Fig. 2).
Environmental response, memory and forgetting in plants
We study how plants memorize environmental temperature and light conditions and reveal the molecular mechanisms that confer the plasticity and robustness of the cascades under various environmental stimuli. These studies will serve as a basis of plant growth optimization for improved crop plant yields (Fig.3).
Epigenetic regulation in sexual reproduction
Heterosis or hybrid vigor is the increased function of any biological quality in a hybrid offspring. We study the epigenetic mechanisms of the heterosis using Arabidopsis accessions. We also study epigenetic-mediated genomic imprinting and self-incompatibility.
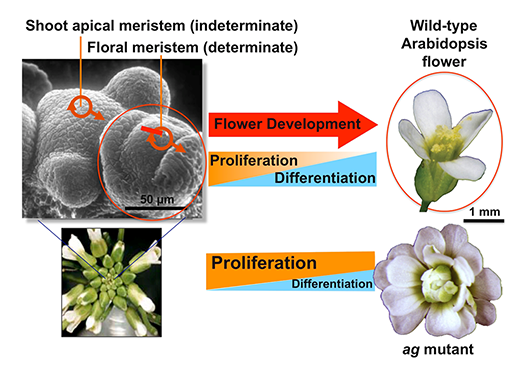
In flower development, the stem cell activities in the floral meristem are terminated (determinate), while the shoot apical meristem continues to grow.
By revealing the mechanisms of floral stem cell regulation and environmental responses, we will develop a molecular basis for plant growth optimization for higher crop yield.
References
- Shirakawa et al., Nature Plants, doi.org/10.1038/s41477-025-01921-1, 2025
- Otsuka et al., Commun. Biol., 8, 108, 2025
- Wang et al., Plant Cell, 36, 5004–5022, 2024
- Wang et al., eLIFE doi.org/10.7554/eLife.100905.1, 2024
- Kobayashi et al., J of Biotech., 392, 103, 2024
- Furuta et al., Nature Commun., 15, 1098, 2024
- Wang et al., Inter J. of Mol Sci., 24, 13297, 2023
- Pelayo et al., Plant Cell, 35, 2821-2847, 2023
- Maruoka et al., Front Plant Sci., 13, 837831, 2022
- Wang et al., Inter J. of Mol Sci., 23, 3864, 2022
- Shirakawa et al., Front. Plant Sci., 12, 829541, 2022
- Yamaguchi et al., Nature Commun., 12, 3480, 2021
- Pelayo et al., Current Opinion Plant Biol., 61, 1020009, 2021
- Shirakawa et al., Front. Plant Sci., 12, 634068, 2021
- Wu et al., Front. Plant Sci., 11, 596835, 2020
- Wang et al., Front. Plant Sci., 11, 600726, 2020

 NAIST Edge BIO(
NAIST Edge BIO(