Applied Stress Microbiology



- Professor
- TAKAGI Hiroshi
- Associate Professor
- KIMATA Yukio, WATANABE Daisuke
- Assistant Professor
- NASUNO Ryo, NISHIMURA Akira, MOROZUMI Yuichi, NAKASE Yukiko, ISOGAI Shota
Outline of Research and Education
Our research involves “Applied Molecular Microbiology”.Our laboratory aims at basic studies in microbial science, particularly cellular response and adaptation to environmental stresses, and its practical applications in new biotechnology. To fully understand microbial cell functions under stress conditions, we analyze and improve various mechanisms of microorganisms from molecular, metabolic and cellular aspects. Our novel findings from our basic studies may be applied to the molecular breeding of useful microbes (yeasts, bacteria), the production of valuable biomaterials (enzymes, amino acids) and the development of promising technologies to solve environmental issues (bioethanol, etc.).
To achieve these goals, we employ research methods in screening, genetics, biochemistry, molecular biology, enzymology, cell biology, post-genomic analysis, protein engineering, metabolic engineering and cell engineering based on the recent knowledge.
Major Research Topics
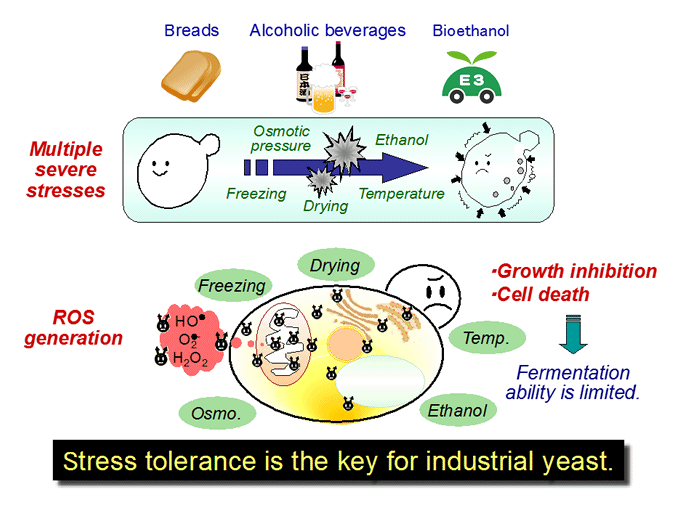
Stress response and tolerance in yeast Saccharomyces cerevisiae (Figs.1, 2, 3, 4)
We are interested in cellular response and adaptation to environmental stresses in the yeast Saccharomyces cerevisiae, which is an important microorganism as a model for higher eukaryotes. Yeast is also a useful microbe in the fermentation industry for the production of breads, alcoholic beverages and bioethanol. During fermentation, yeast cells are exposed to various stresses, including ethanol, high temperature, desiccation and osmotic pressure. Such stresses induce protein denaturation, reactive oxygen species generation, and lead to growth inhibition or cell death. In terms of application, stress tolerance is the key for yeast cells. We analyze the novel stress-tolerant mechanisms found in yeast listed below.
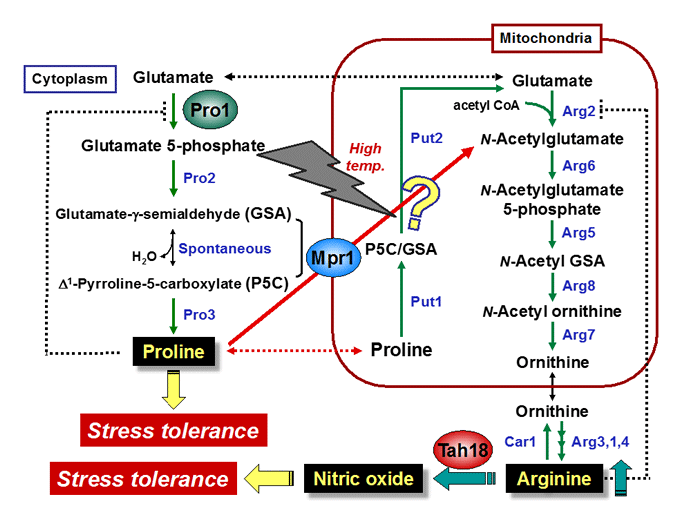
Proline: physiological functions, metabolic regulation, transport mechanisms
In response to osmotic stress, proline is accumulated in many bacterial and plant cells as an osmoprotectant. Previously, we found that proline is a very good cryoprotectant to yeast cells. We currently study the physiological functions and the transport mechanism of proline, and the functions of key enzymes in proline metabolism, γ-glutamyl kinase and proline oxidase.
N-Acetyltransferase Mpr1: arginine biosynthesis, antioxidative mechanisms
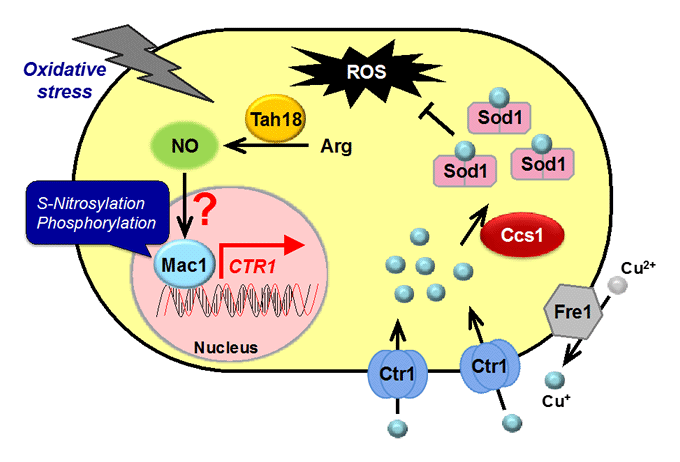
The Mpr1 gene found in S. cerevisiae Σ1278 encodes a novel N-acetyltransferase that detoxifies the proline analogue azetidine-2-carboxylate. We found that Mpr1 is involved in a novel pathway of arginine biosynthesis and protects yeast cells by regulating ROS levels under oxidative stress conditions. We will clarify the mechanism and physiological role of Mpr1-mediated arginine biosynthesis in yeast.
Nitric oxide (NO): synthetic mechanism, physiological roles
Nitric oxide (NO) produced by NO synthase (NOS) from arginine is a signaling molecule participates in many biological processes in mammals, but is poorly understood in yeast due to the lack of mammalian NOS orthologues. We recently discovered that NO is generated through the flavoprotein Tah18-dependent NOS-like activity and confers oxidative stress tolerance on yeast cells. We now analyze the synthetic mechanism and physiological roles of NO in yeast.
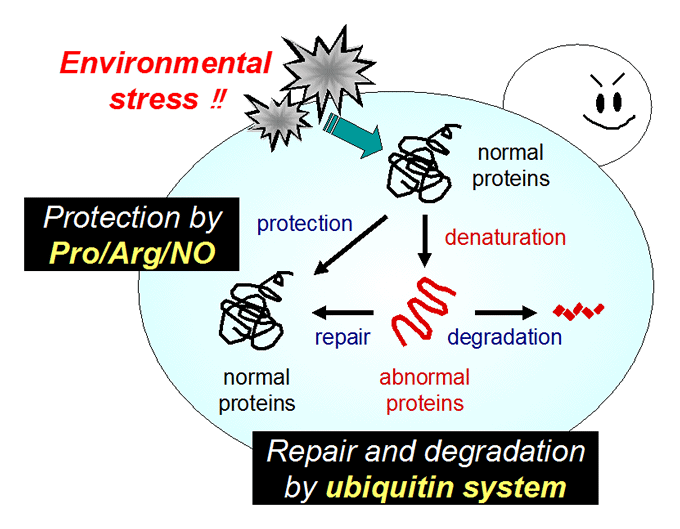
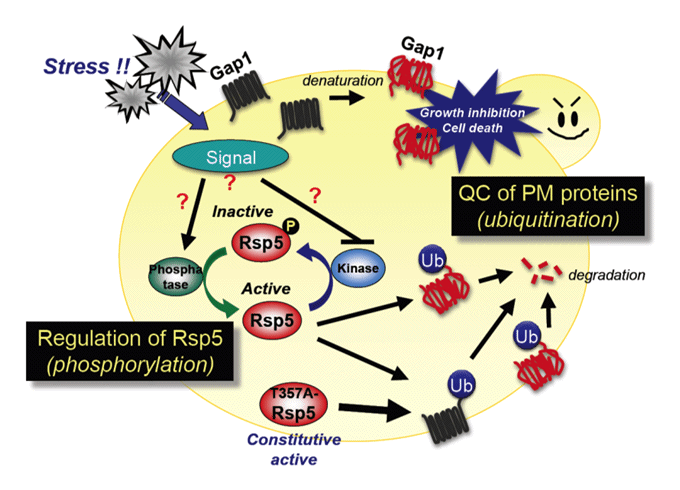
Ubiquitin (Ub) system: plasma membrane protein quality control, Ub ligase Rsp5 regulation.
We are also studying the ubiquitin (Ub) system involving Rsp5, an essential E3 ubiquitin ligase that governs many biological events. We showed that Rsp5 plays pivotal roles in both repair and degradation of stress-induced abnormal proteins. In particular, we hypothesize that Rsp5 is involved in selective ubiquitination for quality control of plasma membrane proteins under stress conditions.
Development of industrial yeast based on novel stress-tolerant mechanisms (Fig. 5)
Through our basic research on novel stress-tolerant mechanisms, we construct industrial yeasts with higher fermentation ability under various stress conditions and contribute to yeast-based industries for the effective production of bread dough and alcoholic beverages or breakthrough in bioethanol production.
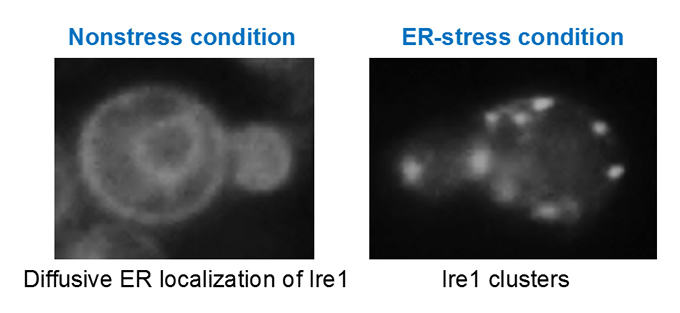
Endoplasmic reticulum (ER) stress and unfolded protein response (UPR) in yeast cells(Fig. 6)
Upon ER stress, eukaryotic cells evoke the UPR to restore dysfunction of the ER. We are pursuing the molecular mechanism by which the ER stress senor protein Ire1 senses ER accumulation of aberrant secretory and transmembrane proteins, clusters and then triggers the UPR in yeast cells.
References
Stress response and tolerance in yeast Saccharomyces cerevisiae
- Eknikom S. et al., Sci. Rep., 12, 4664, 2022
- Nasuno R. et al., Front. Microbiol., 13, 825121, 2022
- Isogai S. et al., Appl. Environ. Microbiol., 87, e0060021, 2021
- Anam K. et al., Sci. Rep., 10, 6015, 2020
- Nishimura A. et al., J. Biol. Chem., 294, 13781-13788, 2019
- Watanabe D. et al., Appl. Environ. Microbiol., 85, e02083-18, 2019
- Takpho N. et al., Metab. Eng., 46, 60-67, 2018
- Yeon J. Y. et al., Sci. Rep., 8, 2377, 2018
- Watcharawipas A. et al., FEMS Yeast Res., 17, fox083, 2017
- Yoshikawa Y. et al., Nitric Oxide-Biol. Chem., 57, 85-91, 2016
- Astuti R. I. et al., Nitric Oxide-Biol. Chem., 52, 29-40, 2016
- Nasuno R. et al., J. Biochem., 159, 271-277, 2016
- Watanabe D. et al., Biochem. Biophys. Res. Commun., 463, 76-81, 2015
- Wijayanti I. et al., J. Biochem., 157, 251-260, 2015
- Nasuno R. et al., PLoS One, 9, e113788, 2014
- Shiga T. et al., Eukaryot. Cell, 13, 1191-1199, 2014
- Nasuno R. et al., Proc. Natl. Acad. Sci. USA, 110, 11821-11826, 2013
- Nomura M. and Takagi H., Proc. Natl. Acad. Sci. USA, 101, 12616-12621, 2004
- Hoshikawa C. et al., Proc. Natl. Acad. Sci. USA, 100, 11505-11510, 2003
Development of industrial yeast based on novel stress-tolerant mechanisms
- Abe T. et al., Front. Genet., 10, 490, doi: 10.3389/fgene.2019.00490, 2019
- Ohashi M. et al., J. Ind. Microbiol. Biotechnol., doi: 10.1007/s10295-019-02177-3, in press
- Watanabe D. et al., Appl. Environ. Microbiol., 84, e00406-18, 2018
- Tsolmonbaatar A. et al., Int. J. Food Microbiol., 238, 233-240, 2016
- Takagi H. et al., J. Biosci. Bioeng., 119, 140-147, 2015
- Sasano Y. et al., Int. J. Food Microbiol., 152, 40-43 , 2012
- Sasano Y. et al., Microb. Cell Fact., 11:40 doi:10.1186/1475-2859-11-40, 2012
Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in yeast cells
- Tran DM. et al., Biosci. Biotech. Biochem., 83, 326-329, 2019
- Mai CT. et al., FEMS Yeast Res., 18, foy016, 2018
- Le QG et al., FEMS Yeast Res., 16, fow049, 2016
- Mathuranyanon R. et al., J. Cell Sci., 128, 1762-1772, 2015
- Ishiwata-Kimata Y. et al., Cell Struct. Funct., 38, 135-143. 2013
- Promlek et al., Mol. Biol. Cell, 22, 3520-3532, 2011
- Kimata et al., J. Cell Biol., 179, 75-86, 2007
