Plant Secondary Metabolism

- Professor
- TOHGE Takayuki

- Assistant Professor
- KOMAKI Shinichiro

- WATANABE Mutsumi

- Labs HP
- https://bsw3.naist.jp/tohge/top_en.html
Outline of Research and Education
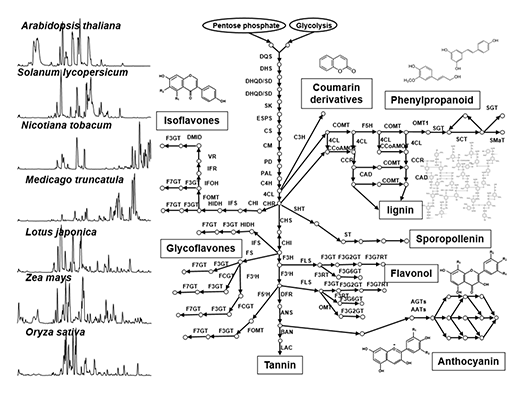
Plant secondary metabolism (also called “specialized metabolism”) produces compounds having several bioactivities such as resistance factors against various environmental stresses in plants, as well as health benefits for humans. Secondary metabolites are widely diversified in their chemical structures in nature (Fig. 1), since plants have adapted to environmental niches during long evolutionary periods using varied strategies such as gene duplication and convergent evolution of some key genes, which contributes to chemical diversity. Our laboratory focuses on model plants, crop species and medicinal plants for i) the analysis of the natural diversity of secondary metabolites, and ii) the functional genomics approach by translational analysis of omics studies (genomics, transcriptomics and mass spectrometry-based metabolomics). The specific goal is identifying key factors of natural chemical diversity and regulatory roles in plant secondary metabolism to enable the metabolic engineering of beneficial compounds.
Major Research Topics
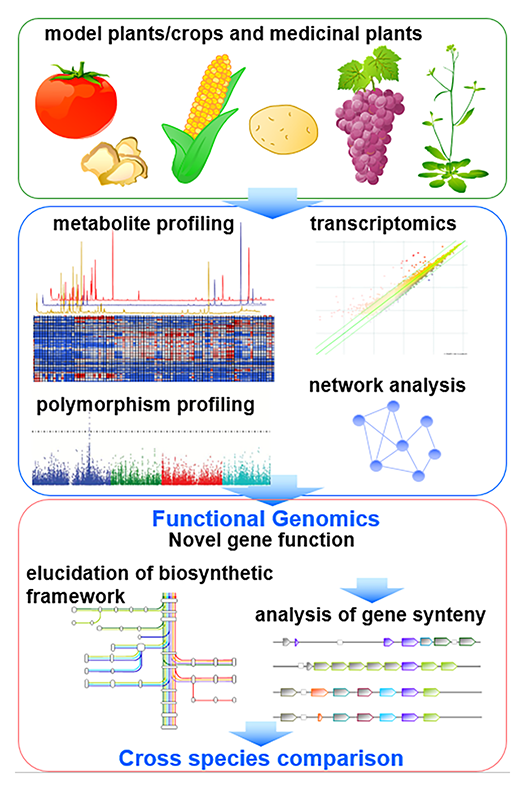
Functional genomics approach by omics-based translational analysis
After completion of full-genome sequencing of huge array of plant species, the complete biosynthetic framework of each plant species still needs to be elucidated, since genome information is not sufficient to compute the size and framework of plant metabolism. We therefore perform metabolomic analysis to screen qualitative differences of metabolite levels between different species, tissues and natural mutants for refinement of recent models of biosynthetic framework (Fig. 2).After illustration of metabolic framework, genome and transcriptome data, as well as genome-wide resources such as quantitative trait locus (QTL) lines and wild accessions for genome-wide association studies (GWAS), are employed for translational analysis. We focus on the discovery of key genes involved in the creation of chemical diversity, and production of beneficial compounds.
Cross species comparison of the neo-functionalized genomic region
The range of genetics-based strategies for characterization of key genes described above provide several genes and genomic regions involved in neo-functionalization of plant secondary metabolism. “Neo-functionalization”, which produces a totally new function after a gene duplication, is a key factor of functional gene divergence. We therefore focus on the species-specific duplicated genes in these key genome synteny regions in order to discover new functional genes in plant secondary metabolism.
Regulation of metabolic networks during nutritional stresses
Nutrient deficiency in soil causes severe reduction in growth with low yields and crop quality. We investigate metabolic and gene expression changes of plants grown under nutrient deprivation stress. This study aims to: i) make an index of time-dependent metabolic changes, ii) evaluate the robustness of metabolic networks, and iii) find species-conserved metabolic makers for the effective breeding of plants having high nutrient-use efficiency or tolerance to nutritional stress.
References
- Watanabe et al., Plant Cell, 20, 2484-2496, 2008.
- Djamei et al., Nature., 478, 395-398, 2011
- Wang et al., Nat Biotech., 32, 1158-1165, 2014
- Bolger et al., Nat Genet., 46, 1034-1038, 2014
- Tohge et al., Plant J., 83, 686-704, 2015
- Aarabi et al., Sci Adv.,2, e1601087, 2016
- Tohge et al., Nat Commun., 7, 12399, 2016
- Dong et al., Nat Commun., 8, 1174 2017
- Peng et al., Nat Commun., 8, 1975, 2017
- Ferrari et al., Nat Commun., 10, 737, 2019
- Perez de Souza et al., Plant Physiol., 182, 857-869, 2019
- Saigo et al., Curr Opin Plant Biol., 55, 93-99, 2020
- Tohge et al., Mol Plant, 13,1-20, 2020
- Calumpang et al., Metabolites, 10, 209, 2020

 NAIST Edge BIO
NAIST Edge BIO