RNA Molecular Medicine

- Professor
- OKAMURA Katsutomo

- Assistant Professor
- SHIIMORI Masami

- Labs HP
- https://bsw3.naist.jp/okamura/
Outline of Research and Education
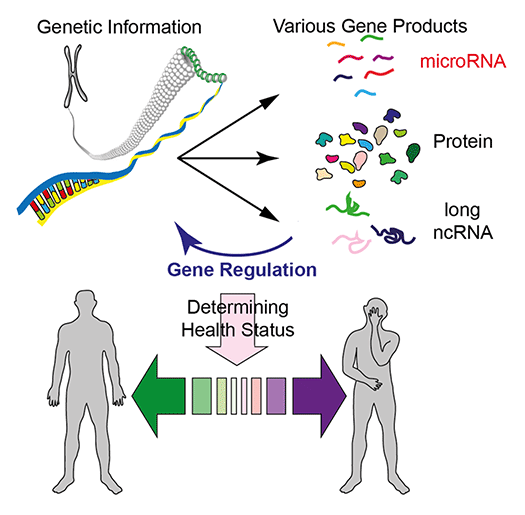
Advances in genomics technologies have transformed research and development strategies in biology and biomedicine, allowing us to access genetic information encoded in our DNA (Fig 1). Our laboratory is interested in understanding how individual genes form large regulatory networks to control biological processes. In particular, we study how regulatory non-coding RNAs including microRNAs (miRNAs) contribute to gene regulation and how their misregulation leads to human health problems.
Research in our laboratory relies on a combination of traditional and modern techniques including biochemistry, genetics and computational biology. Students are expected to learn how to carefully interpret analysis results and develop strategies to answer biological questions by exploiting utilizing technologies or devising new techniques.
Major Research Topics
How is expression of miRNAs controlled?
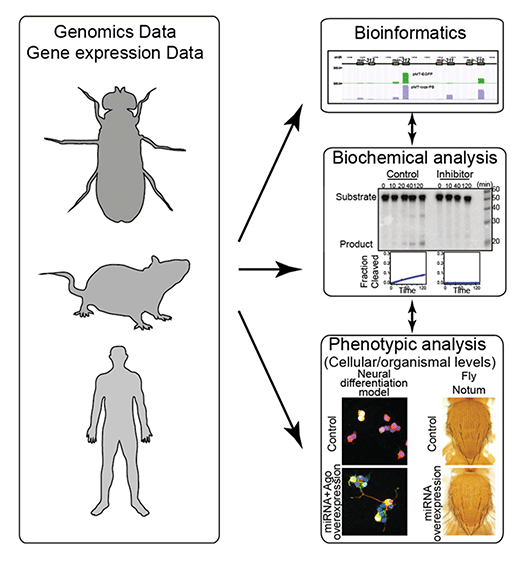
We have witnessed a paradigm shift in the research of gene regulation, and the importance of post-transcriptional regulation of protein-coding genes has now been broadly recognized. Expression of miRNAs should also be regulated at multiple levels (Fig 2). Precise regulation of miRNA levels is important because misregulation of miRNAs often results in human disease. We study how miRNA levels are controlled under healthy and diseased conditions using genomics and biochemical techniques, and examine their biological significance at the cellular and organismal levels (Fig 3).
Why are there many ways to produce miRNAs?
We discovered novel mechanisms of miRNA processing that use machineries known to produce other RNA families, such as mRNA introns and ribosomal RNAs (Fig 2). This means that RNA processing machineries often have unexpected roles in gene regulation. We study the biological significance of non-canonical roles of various RNA processing pathways.
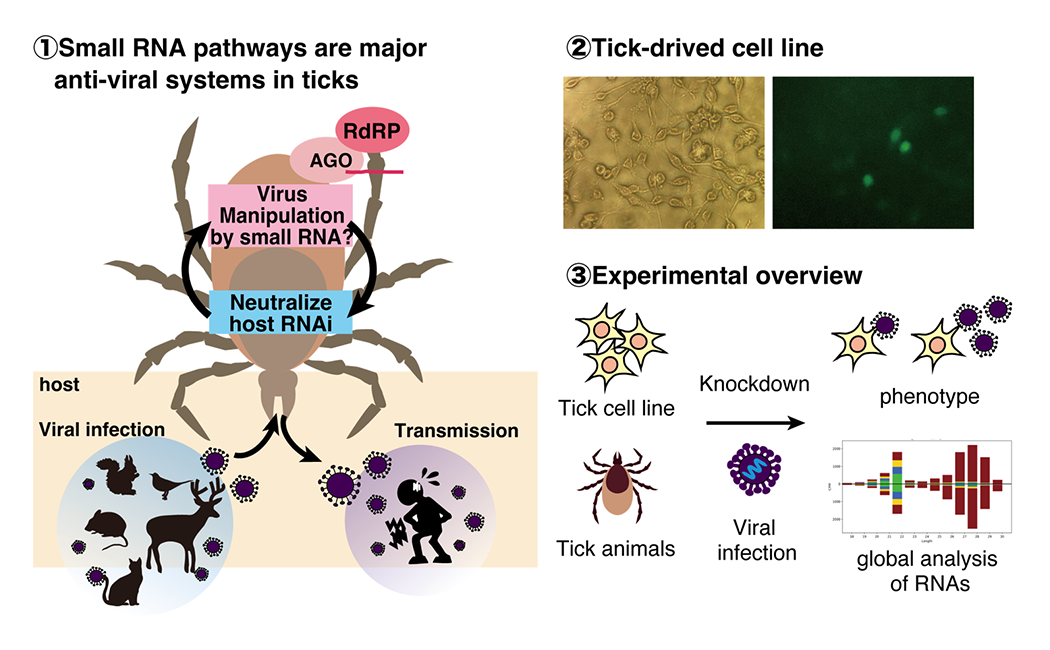
How do tick small RNA pathways regulate viruses?
Small RNA pathways show significant evolutional diversity. Ticks have RNA-dependent RNA polymerases (RdRPs) which are lost in many animals, and we found that ticks have a novel RdRP-dependent small RNA pathway. We study molecular mechanisms underlying interactions of tick small RNA pathways and viruses. Discoveries of new small RNA pathways may pave the way for the development of novel technologies that complement the current CRISPR or RNA interference technologies and viral regulation.
References
- Feng et al. PLoS ONE 18(3): e0281195, 2023
- Shiimori et al., iScience, 6, 102660, 2021
- Goh and Okamura, Nucleic Acids Res., 47, 3101-3116, 2019
- Zhou and Lim et al., eLife, 7, e38389, 2018
- Goh and Okamura, Methods Mol Biol., 1680, 41-63, 2018
- Shiimori et al., Mol Cell., 5, 814-824, 2018
- Lim and Ng et al., Cell Reports, 15, 1795-1808, 2016
- Chak et al., RNA, 21, 375-384, 2015
- Chak and Okamura, Frontiers in Genetics, 5, 172, 2014
- Okamura et al., Genes & Dev, 27, 778-792, 2013

 NAIST Edge BIO
NAIST Edge BIO