Molecular Immunobiology

- Professor
- KAWAI Taro

- Assistant Professor
- ORI Daisuke

- KANO Norisuke

- Labs HP
- https://bsw3.naist.jp/kawai/
Outline of Research and Education
Our body has an immune system to fight against microbial pathogens such as viruses, bacteria, and parasites. There are two arms of the immune system; innate and adaptive immunity. The innate immune system is the first line of host defense that detects invading microbial pathogens and plays a critical role in triggering inflammatory responses as well as shaping adaptive immune responses. In spite of its role in host defense, aberrant activation of innate immune responses is closely associated with exacerbation of inflammatory diseases, autoimmune diseases and cancer. Our aim is to uncover molecular mechanisms that control innate immune responses using tools of molecular and cell biology, bioinformatics and genetically modified mice, and seek a way to control immune diseases.
Major Research Topics
Analysis of innate immune signaling pathways
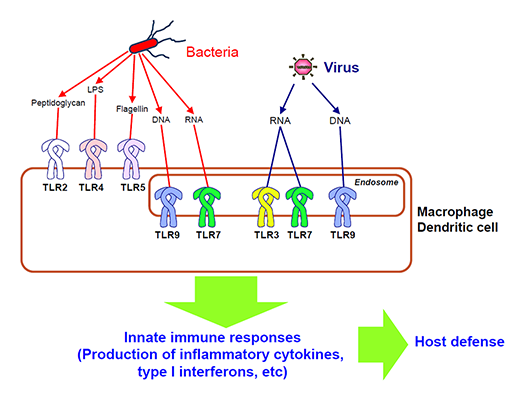
The innate immune system employs germline-encoded pattern-recognition receptors (PRRs) for the initial detection of microbes. PRRs distinguish self from non-self by recognizing microbe-specific molecular signatures known as pathogen-associated molecular patterns (PAMPs), and activate downstream signaling pathways that lead to the induction of innate immune responses by producing inflammatory cytokines, type I interferon (IFN) and other mediators. Mammals have several distinct classes of PRRs including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), Nod-like receptors (NLRs), AIM2-like receptors (ALRs), C-type lectin receptors (CLRs) and intracellular DNA sensors. Among these, TLRs were the first to be identified, and are the best characterized. The TLR family comprises 13 members, which recognize distinct or overlapping PAMPs such as lipid, lipoprotein, protein and nucleic acid (Fig.1). We are focusing on the recognition mechanism of microbial components by PRRs and their signaling pathways, and understanding their roles in immune responses.
Analysis of RLRs
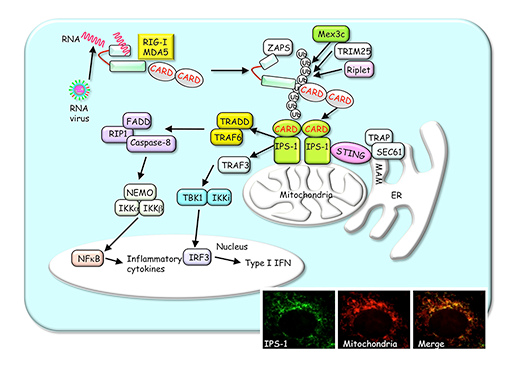
RLRs such as RIG-I and MDA5 are cytoplasmic RNA helicases that detect infection of RNA viruses. Upon detection of RNA virus, RLRs trigger intracellular signaling pathways by recruiting a mitochondria-localized adapter IPS-1, which further activates the transcription factors NF-kB and IRF3 that control expression of antiviral genes, including IFN and inflammatory cytokines (Fig.2). We seek to understand molecular mechanisms underlying RLRs-mediated antiviral innate immune responses.
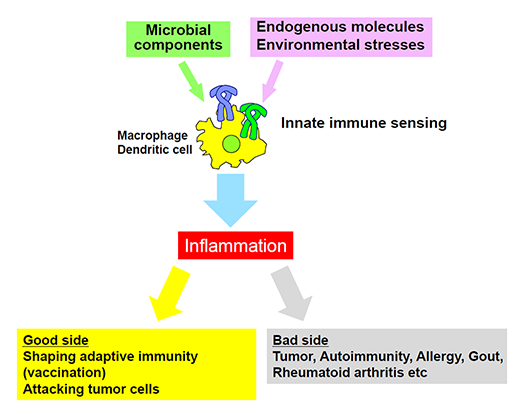
Analysis of sensing mechanisms of endogenous molecules by PRRs (Fig.3)
Recent evidence has shown that innate immunity can react with endogenous molecules derived from necrotic cell death and this reaction is associated with inflammatory diseases. In addition, innate immunity also senses environmental factors such as asbestos and pollen, and causes cancer and allergic responses, respectively. We are seeking the recognition mechanisms of these molecules by innate immunity and its role in diseases.
References
- Kano N. et al., Int Immunol, 36, 471-481, 2024
- Kawai T. et al., Immunity, 57, 649-673, 2024
- Kawasaki T. et al., Cell Rep, 41, 111828, 2022
- Lian BSX. et al., iScience, 25:104118, 2022
- Hasan MZ. et al., Sci Rep, 11, 16814, 2021
- Sok SPM. et al., Int Immunol, 33, 373-386, 2021
- Zainol MIB . et al., Sci Rep. 9, 20406, 2019
- Putri DDDP et al., J Biol Chem., 294, 8412, 2019
- Sueyoshi T. et al., J Immunol., 200, 3814-3824, 2018
- Murase M. et al., J Immunol., 200, 2798-2808, 2018
- Kawasaki T. et al., EMBO J, 36, 1707-1718, 2017
- Kitai Y. et al., J Immunol, 198, 1649-1659, 2017
- Kawasaki T et al., Cell Host Microbe, 14, 148-155, 2013

 NAIST Edge BIO
NAIST Edge BIO