Functional Genomics and Medicine

- Associate Professor
- ISHIDA Yasumasa

- Assistant Professor
- SHIGEOKA Toshiaki

- Labs HP
- https://bsw3.naist.jp/ishida/?cate=431
Outline of Research and Education
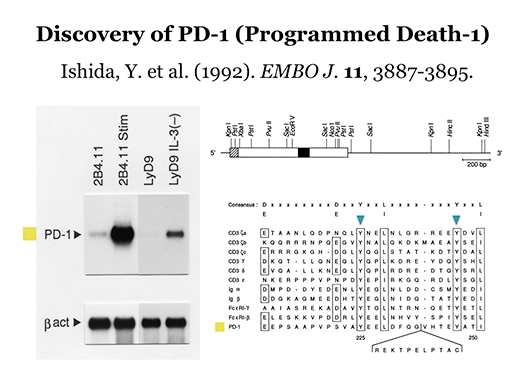
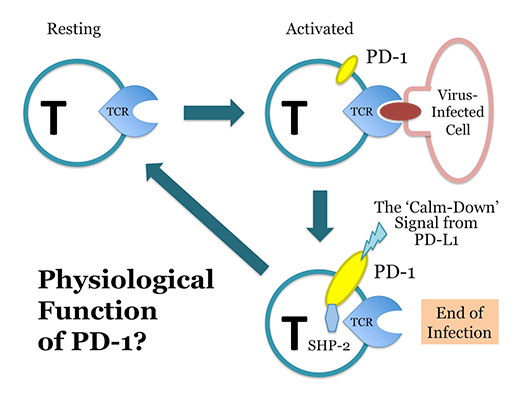
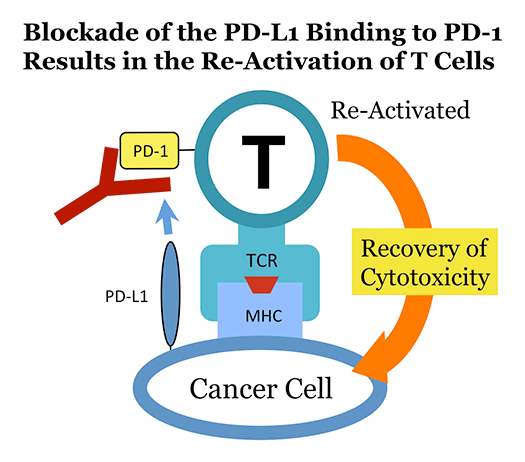
In 1991 at Kyoto University, Ishida et al. discovered a novel gene in a project for the elucidation of the molecular mechanisms involved in the self-nonself discrimination by the immune system, and named it programmed death-1 (PD-1), hoping that it somehow plays a pivotal role when self-reactive (harmful) T lymphocytes (T cells) commit suicide by undergoing apoptosis. PD-1 is a type I transmembrane protein expressed on T cells that are activated by antigenic stimulation. Initially, the physiological function of PD-1 was elusive, but it was shown later that PD-1 downregulates excessive immune reactions. Recently, T. Honjo et al. (Kyoto Univ.) discovered that the cytotoxicity of T cells against some cancer cells can be induced by the antibody-mediated blockade of the above physiological function of PD-1. This anti-cancer strategy is now being widely performed in clinics of many countries, and the Nobel Prize 2018 in physiology and medicine was awarded to T. Honjo (and J.P. Allison). Unfortunately, however, the roles of PD-1 in self-nonself discrimination by the immune system still remain elusive. We conduct our research in the fields of immunology and molecular genetics to identify these roles.
Major Research Topics
Elucidation of the real physiological functions of PD-1
It is very strange that we can cure cancer by blocking the physiological functions of PD-1. What is then PD-1 doing in our body? Is PD-1 on our side (protecting us) or on the side of cancer cells (protecting them)? People believe that PD-1 is a negative regulator of the immune responses, but what kind of signals in the immune system is PD-1 suppressing? (Obviously, PD-1 is not an omnipotent negative regulator in the immune system) To answer these questions, we perform experiments in immunology and molecular biology by using a variety of genetically modified animals (including PD-1 knockouts).
Development of novel strategies in cancer immunotherapy
Cancer immunotherapy based on the blockade of the physiological functions of PD-1 is effective only upon a limited number of cancer patients. For instance, only about 20% of lung-cancer patients and only about 30% of melanoma patients show good responses to such a PD-1-blocking strategy. We try to improve this low efficacy of current cancer immunotherapy by creating a variety of "oncolytic" recombinant retroviruses.
References
- Bai J. et al., Genesis in press, 2020
- Ishida Y., Cells in press, 2020
- Yamanishi A. et al., Nucleic Acids Res., 46, e63, 2018
- Nakamura A. et al., Neurosci. Res., 100, 55-62, 2015
- Shigeoka T. et al., Nucleic Acids Res., 40, 6887-6897, 2012
- Mayasari N. I. et al., Nucleic Acids Res., 40, e97, 2012
- Shigeoka T. et al., Nucleic Acids Res., 33, e20, 2005
- Matsuda E. et al., Proc. Natl. Acad. Sci. USA, 101, 4170-4174, 2004
- Ishida Y. and Leder P., Nucleic Acids Res., 27, e35, 1999
- Ishida Y. et al., EMBO J., 11, 3887-3895, 1992

 NAIST Edge BIO(
NAIST Edge BIO(