Molecular Microbiology and Genetics


- Professor
- INUI Msayuki

- Associate Professor
- KOGURE Takahisa

- Labs HP
- https://www.rite.or.jp/bio/en/naist/
Outline of Research and Education
Global warming resulting from elevated CO2 and global energy supply problems have been in the limelight in recent years. As these problems originate from rapid economic expansion and regional instability in parts of the world, broad knowledge of global economic systems as well as R&D is necessary to solve these problems. Fundamental research employing microbial functions to tackle adverse effects of global climate change and mitigate energy supply problems is carried out in our laboratory.
Major Research Topics
Biorefinery
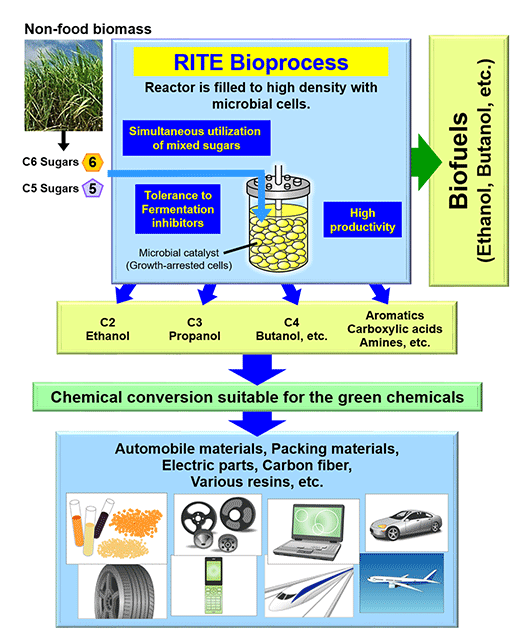
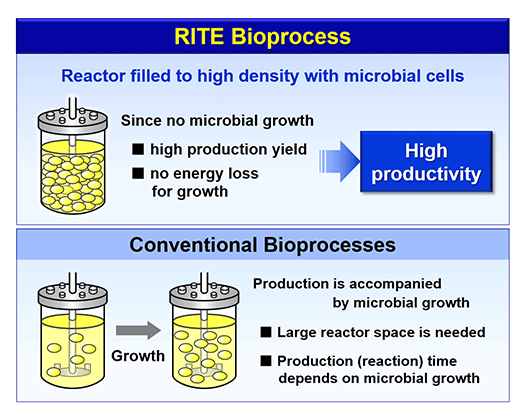
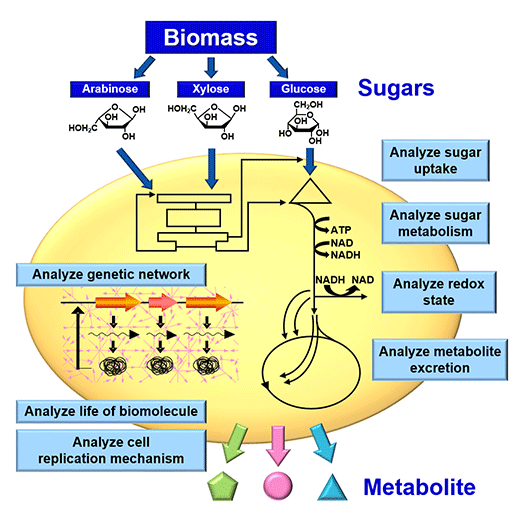
A biorefinery is a concept which describes production of chemicals and fuels from renewable biomass via biological processes. Biorefinery R&D is considered of national strategic importance to the U.S.A. (Fig. 1). A biorefinery can be divided into two processes: a saccharification process to hydrolyze biomass to sugars, and a bioconversion process to produce chemicals and fuels from the sugars. We have pioneered, as a bioconversion technology to produce chemicals and fuels, a highly-efficient “growth-arrested bioprocess” based on a novel concept (Fig.2). It is based on Corynebacteria that are widely used in industrial amino acid production. Key to the high efficiency is the productivity of artificially growth-arrested microbial cells, cells with which we are evaluating production of organic acids and biofuels. To efficiently produce these products, the cells are tailored for the production of a particular product using post genome technologies like transcriptomics, proteomics and metabolome analyses (Fig.3).
Bioenergy and green chemicals production
Having established the fundamental technology to produce bioethanol from non-food biomass, we are now partnering with the automobile and petrochemical industries to explore commercial application. We have also developed the platform technology to produce biobutanol, the expected next-generation biofuel, as well as a variety of green chemicals such as organic acids, alcohols and aromatic compounds from which diverse polymer raw materials used in various industries are produced.
References
- Shimizu T. et al., Appl Microbiol Biotechnol, 108, 1-11, 2024
- Nakamichi Y. et al., Acta Crystallogr D Struct Biol, D79, 895-908, 2023
- Teramoto H. et al., Int J Hydrog Energy, 47, 29219-29229, 2022
- Teramoto H. et al., Int J Hydrog Energy, 47, 22010-22021, 2022
- Shimizu T. et al., Appl Environ Microbiol, 88, e0050722, 2022
- Toyoda K. et al., Microorganisms, 10, 1002, 2022
- Tanaka Y. et al., J Bacteriol, 204, e0005322, 2022
- Nakazawa S. et al., ACS Synth Biol, 10, 2308-2317, 2021
- Toyoda K. et al., Microorganisms, 9, 550, 2021
- Jojima T. et al., Biotechnol Biofuels, 14, 45, 2021
- Kogure T. et al., Metab Eng, 65, 232-242, 2021
- Uchikura H. et al., Appl Microbiol Biotechnol, 104, 6719-6729, 2020
- Hasegawa S. et al., Metab Eng, 59, 24-35, 2020
- Shimizu T. et al., Appl Microbiol Biotechnol, 103, 9739-9749, 2019
- Han S.O. et al., Biotechnol J, 14, e1900160, 2019
- Tsuge Y. et al., Appl Microbiol Biotechnol, 103, 3381-3391, 2019
- Oide S. et al., Enzyme Microb Technol, 125, 13-20, 2019
- Shimizu T. et al., Appl Environ Microbiol, 85, e01873-18, 2019
- Tsuge Y. et al., J Biosci Bioeng, 127, 288-293, 2019
- Kogure T. et al., Appl Microbiol Biotechnol, 102, 8685-8705, 2018