Research Projects
2. Metabolic regulatory mechanisms and physiological roles of functional amino acids and their applications in yeast (Functional amino acid engineering)
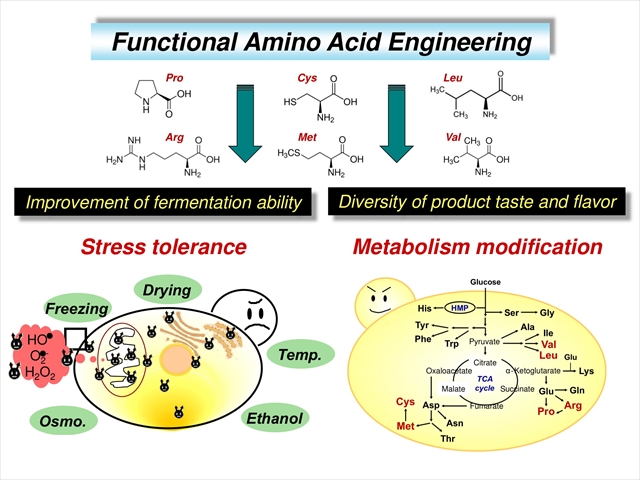
In yeast, amino acid metabolism and its regulatory mechanisms vary under different growth environments by regulating anabolic and catabolic processes, including uptake and export, and the metabolic styles form a complicated but robust network. There is also crosstalk with various metabolic pathways, products and signal molecules. The elucidation of metabolic regulatory mechanisms and physiological roles is important fundamental research for understanding life phenomenon. In terms of industrial application, the control of amino acid composition and content is expected to contribute to an improvement in productivity, and to add to the value of fermented foods, alcoholic beverages, bioethanol, and other valuable compounds (proteins and amino acids, etc.). We developed a new method “functional amino acid engineering” for constructing yeast strains with high functionality focused on the metabolic regulatory mechanisms and physiological roles of ‘functional amino acids’, found in yeast.

- A. Nishimura, R. Tanahashi, K. Nakagami, Y. Morioka and H. Takagi: Identification of an arginine transporter in Candida glabrata. J. Gen. Appl. Microbiol., in press. DOI: 10.2323/jgam.2023.03.003
- S. Isogai, A. Nishimura, N. Murakami, N. Hotta, A. Kotaka, Y. Toyokawa, H. Ishida and H. Takagi: Improvement of valine and isobutanol production in sake yeast by Ala31Thr substitution in the regulatory subunit of acetohydroxy acid synthase. FEMS Yeast Res., 23, foad012 (2023).
- A. Nishimura, R. Tanahashi, T. Oi, K. Kan and H. Takagi: Plasmid-free CRISPR/Cas9 genome editing in Saccharomyces cerevisiae. Biosci. Biotech. Biochem., 87, 458-462 (2023).
- R. Tanahashi, A. Nishimura, F. Morita, H. Nakazawa, A. Taniguchi, K. Ichikawa, K. Nakagami, K. Boundy-Mills and H. Takagi: The arginine transporter Can1 acts as a transceptor for regulation of proline utilization in the yeast Saccharomyces cerevisiae. Yeast, 40, 333-348 (2023).
- R. Tanahashi, A. Nishimura, M. Nguyen, I. Sitepu, G. Fox, K. Boundy-Mills and H. Takagi: Large-scale screening of yeast strains that can utilize proline. Biosci. Biotech. Biochem., 87, 358-362 (2023) DOI: 10.1093/bbb/zbac202
- M. Tsukahara, S. Isogai, H. Azuma, K. Tsukahara, Y. Toyokawa and H. Takagi: Characterization of a new Saccharomyces cerevisiae isolated from banana stems and its mutant with l-leucine accumulation for awamori brewing. Biosci. Biotech. Biochem., 87, 240-244 (2023). DOI: 10.1093/bbb/zbac185
- A. Nishimura, K. Ichikawa, H. Nakazawa, R. Tanahashi, F. Morita, Ir. Sitepu, K. Boundy-Mills, G. Fox and H. Takagi: The Cdc25/Ras/cAMP-dependent protein kinase A signaling pathway regulates proline utilization in wine yeast Saccharomyces cerevisiae under a wine fermentation model. Biosci. Biotech. Biochem., 86, 1300-1307 (2022).
- A. Nishimura, K. Nakagami, K. Kan, F. Morita and H. Takagi: Arginine inhibits Saccharomyces cerevisiae biofilm formation by inducing endocytosis of the arginine transporter Can1. Biosci. Biotech. Biochem., 86, 1318-1326 (2022).
- J. Koonthongkaew, N. Ploysongsri, Y. Toyokawa, V. Ruangpornvisuti and H. Takagi: Improvement of fusel alcohol production by engineering of the yeast branched-chain amino acid aminotransaminase. Applied and Environmental Microbiology, 10, e00822-22 (2022).
- M. Ohashi, S. Isogai and H. Takagi: Functional analysis of feedback inhibition-insensitive variants of N-acetyl glutamate kinase found in sake yeast mutants with ornithine overproduction. Microbiol. Spectr., 10, e00822-22 (2022).
- S. Isogai, A. Nishimura, A. Kotaka, N. Murakami, N. Hotta, H. Ishida and H. Takagi: High-level production of isoleucine and fusel alcohol by expression of the feedback inhibition-insensitive threonine deaminase in Saccharomyces cerevisiae. Appl. Environ. Microbiol., 88, e02130-21 (2022).
- A. Nishimura, S. Isogai, N. Murakami, N. Hotta, A. Kotaka, K. Matsumura, Y. Hata, H. Ishida and H. Takagi*: Isolation and analysis of a sake yeast mutant with phenylalanine accumulation. J. Ind. Microbiol. Biotechnol., in press. DOI: 10.1093/jimb/kuab085
- Y. Toyokawa†, J. Koonthongkaew† and H. Takagi*: An overview of branched-chain amino acid aminotransferases: functional differences between mitochondrial and cytosolic isozymes in yeast and human. Appl. Microbiol. Biotechnol., 105, 8059-8072 (2021).
- A. Nishimura, Y. Takasaki, S. Isogai, Y. Toyokawa, R. Tanahashi and H. Takagi: Role of Gln79 in feedback inhibition of the yeast γ-glutamyl kinase by proline. Microorganisms, 9(9), 1902 (2021).
- S. Isogai and H. Takagi: Enhancement of lysine biosynthesis confers high-temperature stress tolerance to Escherichia coli cells. Appl. Microbiol. Biotechnol., 105, 6899–6908 (2021).
- S. Isogai, T. Matsushita, H. Imanishi, J. Koonthongkaew, Y. Toyokawa, A. Nishimura, X. Yi, R. Kazlauskas and H. Takagi: High-level production of lysine in the yeast Saccharomyces cerevisiae by rational design of homocitrate synthase. Appl. Environ. Microbiol., 87(15), e0060021 (2021)..
- M. Ohashi, R. Nasuno, S. Isogai and H. Takagi: High-level production of ornithine by expression of the feedback inhibition-insensitive N-acetyl glutamate kinase in the sake yeast Saccharomyces cerevisiae. Metab. Eng., 62, 1-9 (2020). DOI: 10.1016/j.ymben.2020.08.005
- N. Murakami, A. Kotaka, S. Isogai, K. Ashida, A. Nishimura, K. Matsumura, Y. Hata, H. Ishida and H. Takagi: Effects of a novel variant of the yeast γ-glutamyl kinase Pro1 on its enzymatic activity and sake brewing. J. Ind. Microbiol. Biotechnol., 47, 715-723 (2020) DOI: 10.1007/s10295-020-02297-1
- J. Koonthongkaew, Y. Toyokawa, M. Ohashi, C. Large, M. Dunham and H. Takagi: Effect of the Ala234Asp substitution in the mitochondrial branched-chain amino acidaminotransferase Bat1 on the production of BCAAs and fusel alcohols in yeast. Appl. Microbiol. Biotechnol., 104, 7915-7925 (2020).
- N. S. M. Nanyan and H. Takagi: Proline homeostasis in Saccharomyces cerevisiae: How does the stress-responsive transcription factor Msn2 play a role? Front. Genet., 11, 438 (2020). DOI: 10.3389/fgene.2020.00438.
- N. S. M. Nanyan, D. Watanabe, Y. Sugimoto and H. Takagi: Effect of the deubiquitination enzyme gene UBP6 on the stress-responsive transcription factor Msn2-mediated control of the amino acid permease Gnp1 in yeast. J. Biosci. Bioeng., 129, 423-427 (2020). doi: 10.1016/j.jbiosc.2019.10.002.
- N. S. M. Nanyan, D. Watanabe, Y. Sugimoto and H. Takagi: Involvement of the stress-responsive transcription factor gene MSN2 in the control of amino acid uptake in Saccharomyces cerevisiae. FEMS Yeast Res., 19, foz052 (2019).
- T. Abe, Y. Toyokawa, Y. Sugimoto, H. Azuma, K. Tsukahara, R. Nasuno, D. Watanabe, M. Tsukahara* and H. Takagi*: Characterization of a new Saccharomyces cerevisiae isolated from hibiscus flower and its mutant with L-leucine accumulation for awamori brewing. *Co-corrensponding authors. Front. Genet., 10, 490 (2019). doi: 10.3389/fgene.2019.00490.
- H. Takagi: Metabolic regulatory mechanisms and physiological roles of functional amino acids and their applications in yeast. Biosci. Biotech. Biochem., 83, 1449-1462 (2019).
- N. Takpho, D. Watanabe and H. Takagi*: Valine biosynthesis in Saccharomyces cerevisiae is regulated by the mitochondrial branched-chain amino acid aminotransferase Bat1. Microbial Cell, 5, 293-299 (2018).
- N. Takpho, D. Watanabe and H. Takagi*: High-level production of valine by expression of the feedback inhibition-insensitive acetohydroxyacid synthase in Saccharomyces cerevisiae. Metab. Eng., 46, 60-67 (2018).
- J. Y. Yeon, S. J. Yoo, H. Takagi* and H. A. Kang*: A novel mitochondrial serine O-acetyltransferase, OpSAT1, plays a critical role in sulfur metabolism in the thermotolerant methylotrophic yeast Ogataea parapolymorpha. *Co-corrensponding authors. Sci. Rep., 8:2377 DOI: 10.1038/s41598-018-20630-8 (2018).
- H. Takagi, K. Hashida, D. Watanabe, R. Nasuno, M. Ohashi, T. Iha, M. Nezuo, and M. Tsukahara: Isolation and characterization of awamori yeast mutants with L-leucine accumulation that overproduce isoamyl alcohol. J. Biosci. Bioeng., 119, 140-147 (2015).
