Research outcomes
Decoding the Rice Root Microbiome in a High-Yield, Fertilizer- and Pesticide-Free Field: A Four-Year Study with Machine Learning Reveals Beneficial Bacteria for Sustainable Cultivation
Decoding the Rice Root Microbiome in a High-Yield, Fertilizer- and Pesticide-Free Field: A Four-Year Study with Machine Learning Reveals Beneficial Bacteria for Sustainable Cultivation
Summary:
A new study published in Plant and Cell Physiology sheds light on how rice plants grown without chemical fertilizers or pesticides develop and manage their root-associated microbial communities during their adaptation to nutrient-deficient soils. The research offers key insights that may help promote sustainable agriculture by harnessing root-inhabiting microbes to support crop health and productivity.
A research team led by Drs. Asahi Adachi, John Jewish Dominguez, and Professor Yusuke Saijo at the Nara Institute of Science and Technology (President: Kazuhiro Shiozaki), in collaboration with The University of Tokyo, Tokyo Institute of Technology, Nagoya University and Tohoku University, has uncovered a diverse group of root-inhabiting bacteria - both known and previously uncharacterized - that may support rice growth during cultivation without fertilizers. Their findings offer promising directions for developing microbe-based biostimulants to support more sustainable agriculture.
Using 16S rRNA amplicon sequencing1 and machine learning analysis, the team discovered specific bacterial groups that are consistently enriched in the roots of rice plants cultivated in a high-yielding, pesticide-free, and non-fertilized paddy field. This field soil has remained untouched by chemical inputs for over 70 years yet still produces up to 60-70% of the yield of conventionally managed fields.
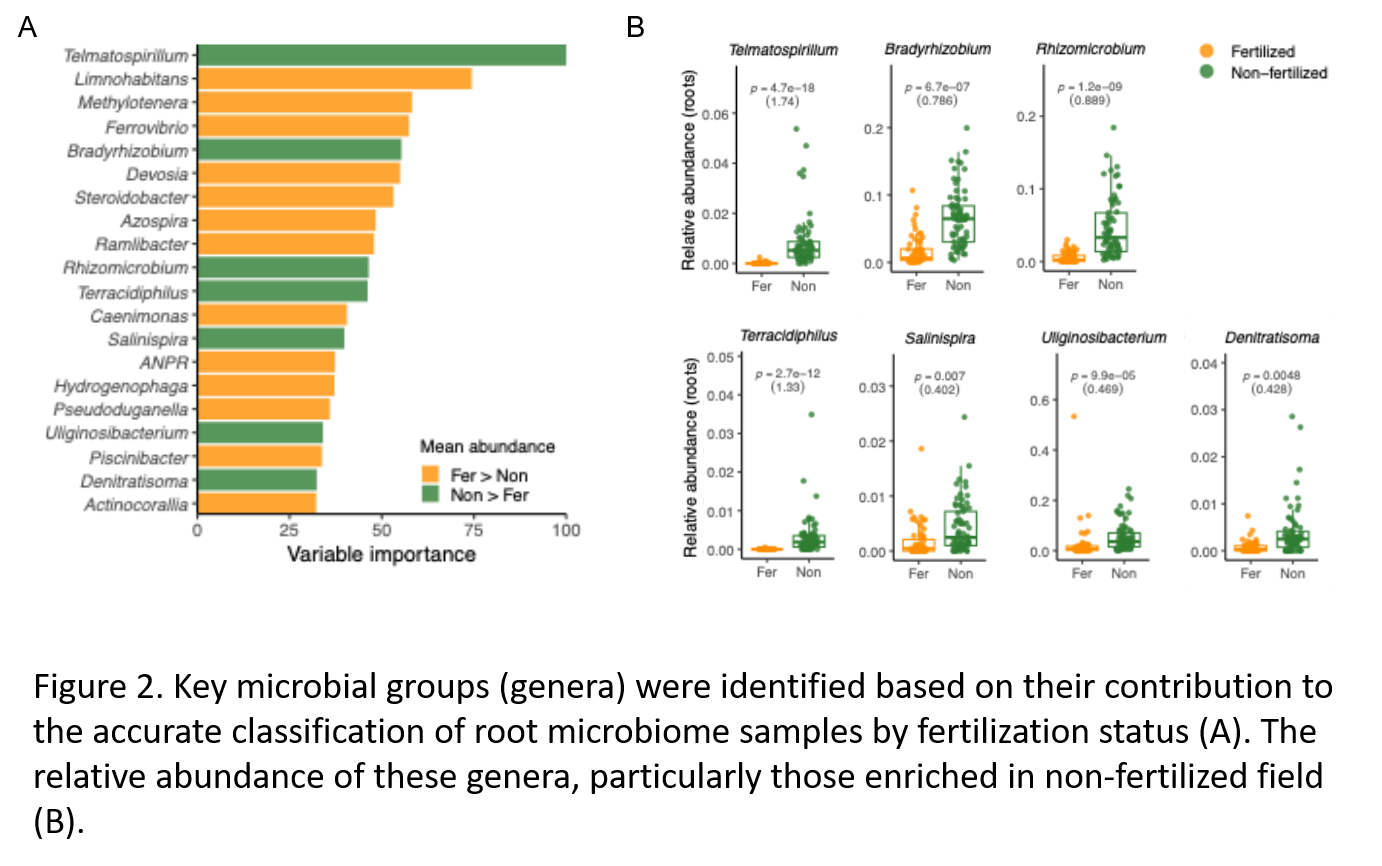
The study monitored rice root microbiomes across four growing seasons and compared microbial communities between the non-fertilized field and a neighboring fertilized field. Among the most striking discoveries was a consistent enrichment of nitrogen-fixing bacteria — including familiar genera like Bradyrhizobium as well as lesser-known taxa such as Telmatospirillum. These bacteria are thought to play a critical role in supplying nitrogen to the plant in the absence of synthetic fertilizers.
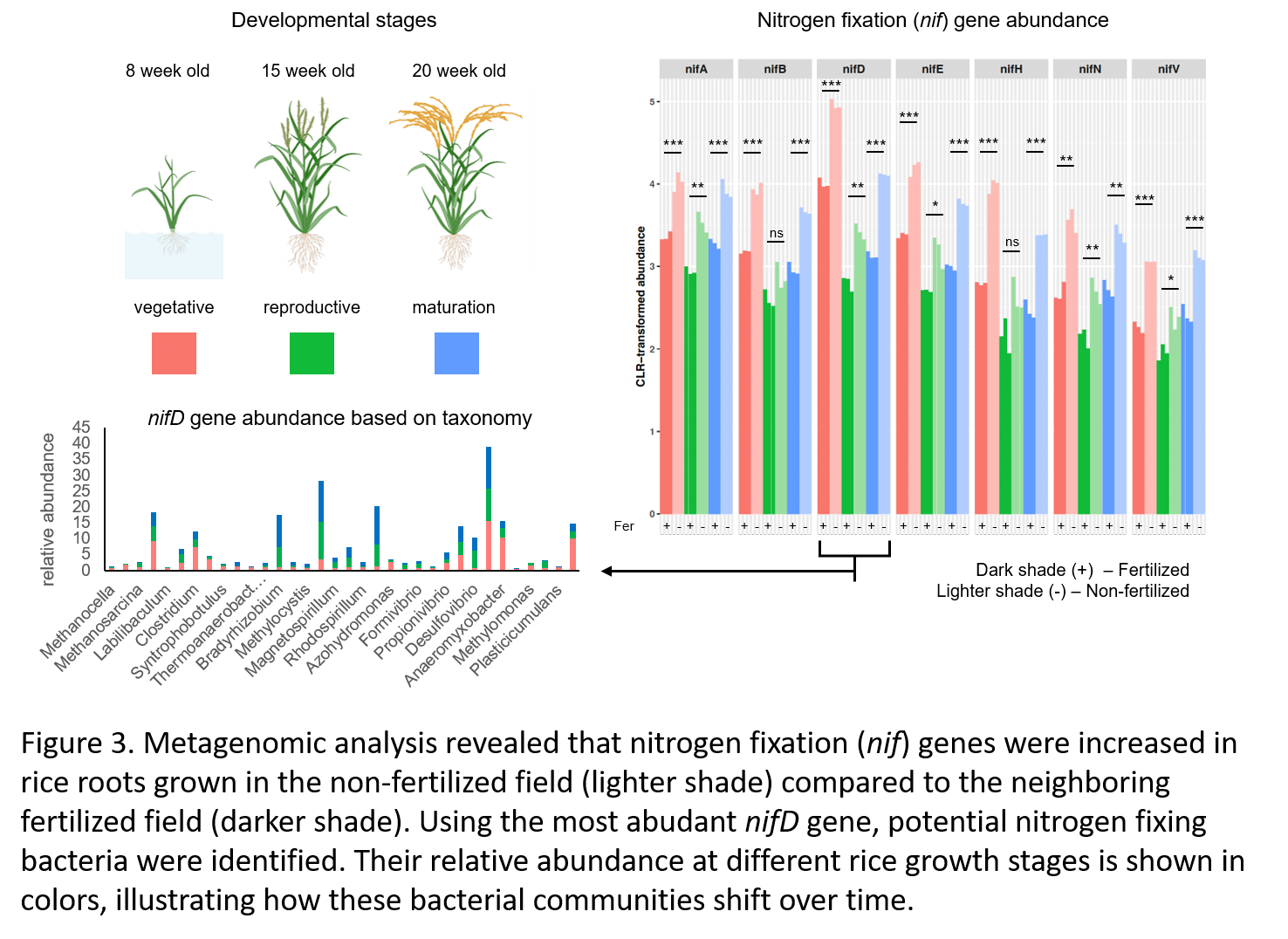
Metagenomic analysis2 further confirmed that genes responsible for nitrogen fixation (nif) were significantly more abundant in the roots of rice grown in the non-fertilized field. Interestingly, the composition of these nitrogen-fixing communities shifted with the plant’s growth stages - from anaerobic bacteria during early vegetative growth to microaerophilic and aerobic bacteria in the maturation phase - likely reflecting changes in soil oxygen availability driven by seasonal variation and cultivation practices such as water drainage.
Background and Purpose:
Plants can adapt to nutrient-poor soils by recruiting beneficial microorganisms, yet the rules governing this recruitment under field conditions remain poorly understood. The unique conditions of the long-term, no-input (NOChI) paddy field provided a rare opportunity to study these interactions. This site, managed without any chemical or organic inputs, serves as a valuable model for investigating how rice maintains productivity through microbiome support under nutrient-limited conditions.
Specifically, this study aimed to (1) characterize root endosphere microbiomes in rice under contrasting fertilization regimes, (2) identify microbial taxa enriched in nutrient-poor soil environments, and (3) examine how plant host factors, such as CCaMK, a master regulator of mycorrhizal symbiosis, influence microbiome assembly.
Study Highlights:
• Fertilization Drives Root Microbiome Structure: The microbiome composition diverged markedly depending on the fertilization regime, while rice cultivar differences had a smaller effect.
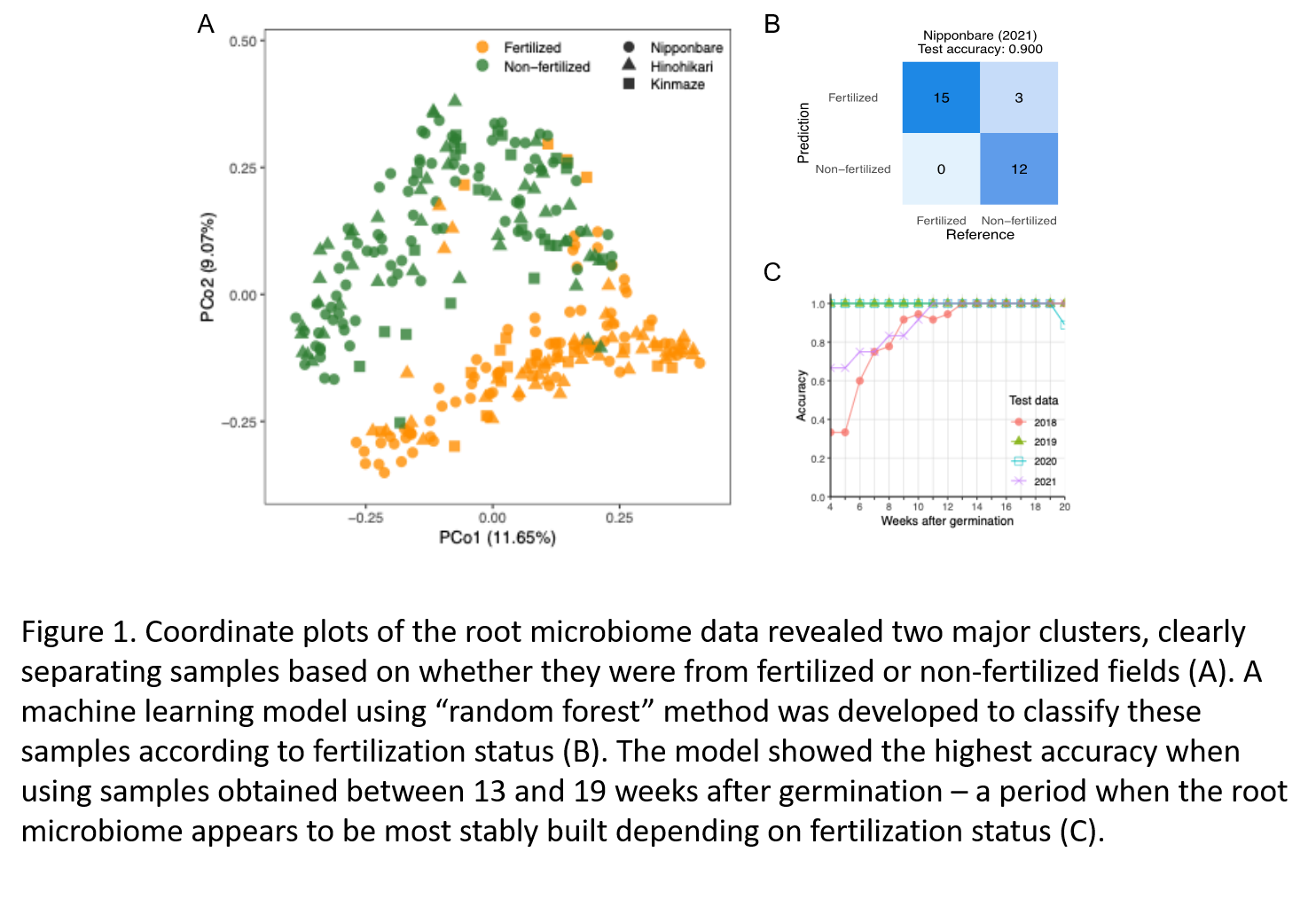
• Machine Learning Prediction Model: A random forest model trained on microbiome data could accurately predict the fertilization status of rice plants, with peak performance observed during mid to late development stages.
• Key Nitrogen-Fixing Taxa Identified: Relative abundance and functional gene analysis revealed microbial genera enriched in paddy rice roots under nutrient-poor conditions, where typical mycorrhizal symbiosis is not well established.
• Dynamic Microbial Shifts: The composition of nitrogen-fixing bacterial populations changed across rice developmental stages, suggesting a close interplay between root microbiome dynamics and cultivation practices.
To explore factors shaping the rice root microbiome, beta-diversity analysis revealed that fertilization regime, rather than rice cultivar or CCaMK symbiosis regulator function, is the primary driver of microbiome structure (Figure 1A). Based on these distinct microbiome profiles (green and orange, in non-fertilized and fertilized fields, respectively), a random forest model trained on samples from 2018–2020 accurately predicted fertilization status in 2021 samples (accuracy = 0.9; Figure 1B), with peak performance observed during mid to late growth stages (13–19 weeks post-germination), indicating a period of microbiome stability (Figure 1C).
Key microbial taxa enriched in the non-fertilized field were identified through machine learning (Figure 2A) and validated by relative abundance and metagenomic analyses (Figure 2B). Nitrogen fixation (nif) genes were consistently more abundant in these samples, with taxonomic profiling of nifD genes revealing rice developmental stage-specific microbial communities - from anaerobes in early growth to microaerophilic and aerobic taxa at maturation (Figure 3). These shifts likely reflect oxygen level changes driven by cultivation practices such as water drainage.
Overall, this study highlights enhanced nitrogen fixation as a key mechanism supporting rice growth in “NOChI” field and identifies novel candidate bacteria, including Telmatospirillum, that may function independently of canonical symbiotic regulators such as CCaMK.
Future Prospects:
This research demonstrates that paddy rice roots host a dynamic microbiome capable of adapting to the absence of chemical fertilizers by enhancing biological nitrogen fixation. Importantly, the ability to predict fertilization status based on microbiome profiles introduces new possibilities in precision agriculture - including assessing the suitability of NOChI-type cultivation, monitoring plant nutrient status, and optimizing fertilizer application for greater efficiency.
Integrating microbiome data with conventional bacterial isolation techniques can accelerate the development of custom-designed microbial consortia - or synthetic communities - as biostimulants for sustainable rice cultivation.
These findings will be published in Plant and Cell Physiology on 9th June, at 9:01 JST.
DOI: 10.1093/pcp/pcaf045
Title: Field Dynamics of the Root Endosphere Microbiome Assembly in Paddy Rice Cultivated under No Fertilizer Input
Authors: Asahi Adachi, John Jewish Dominguez, Yuniar Devi Utami, Masako Fuji, Sumire Kirita, Shunsuke Imai, Takumi Murakami, Yuichi Hongoh, Rina Shinjo, Takehiro Kamiya, Toru Fujiwara, Kiwamu Minamisawa, Naoaki Ono, Shigehiko Kanaya, Yusuke Saijo
116S rRNA amplicon sequencing: A method used to identify and profile microbial communities, particularly bacteria (and also Archaea), by targeting specific regions of the 16S ribosomal RNA gene, a universal genetic marker in prokaryotes.
2Metagenomic sequencing: A technique that analyzes all DNA extracted from samples, enabling the identification and quantification of genes, including those involved in important functions like nitrogen fixation - to assess the overall capabilities of microbial communities.
「EurekAlert!」
URL:https://www.eurekalert.org/news-releases/1090037
Laboratory of Plant Immunity
https://bsw3.naist.jp/eng/courses/courses111.html
https://bsw3.naist.jp/saijo/en/home/
( June 13, 2025 )
