Research outcomes
AI knows where your proteins go
Researchers from Nara Institute of Science and Technology find that machine learning can predict the subcellular locations of functionally related proteins
Facial recognition software can be used to spot a face in a crowd; but what if it could also predict where someone else was in the same crowd? While this may sound like science fiction, researchers from Japan have now shown that artificial intelligence can accomplish something very similar on a cellular level.
In a study published in Frontiers in Cell and Developmental Biology, researchers from Nara Institute of Science and Technology (NAIST) have revealed that a machine learning program can accurately predict the location of proteins related to actin, an important part of the cellular skeleton, based on the location of actin itself.
Actin plays a key role in providing shape and structure to cells, and during cell movement helps form lamellipodia, which are fan-shaped structures that cells use to "walk" forwards. Lamellipodia also contain a host of other proteins that bind to actin to help maintain the fan-like structure and keep the cells moving.
"While artificial intelligence has been used previously to predict the direction of cell migration based on a sequence of images, so far it has not been used to predict protein localization," says lead author of the study, Shiro Suetsugu. This idea came in during the discussion with Yoshinobu Sato at the Data Science Center in NAIST. "We therefore sought to design a machine learning algorithm that can determine where proteins will appear in the cell based on their relationship with other proteins."
To do this, the researchers trained an artificial intelligence system to predict where actin-associated proteins would be in the cell by showing it pictures of cells in which the proteins were labeled with fluorescent markers to show where they were located. Then, they gave the program pictures in which only actin was labeled and asked it to tell them where the associated proteins were.
"When we compared the predicted images to the actual images, there was a considerable degree of similarity," states Suetsugu. "Our program accurately predicted the localization of three actin-associated proteins within lamellipodia; and, in the case of one of these proteins, in other structures within the cell."
On the other hand, when the researchers asked the program to predict where tubulin, which is not directly related to actin, would be in the cell, the program did not perform nearly as well.
"Our findings suggest that machine learning can be used to accurately predict the location of functionally related proteins and describe the physical relationships between them," says Suetsugu.
Given that lamellipodia are not always easy for non-experts to spot, the program developed in this study could be used to quickly and accurately identify these structures from cell images in the future. In addition, this approach could potentially be used as a sort of artificial cell staining method to avoid the limitations of current cell-staining methods.
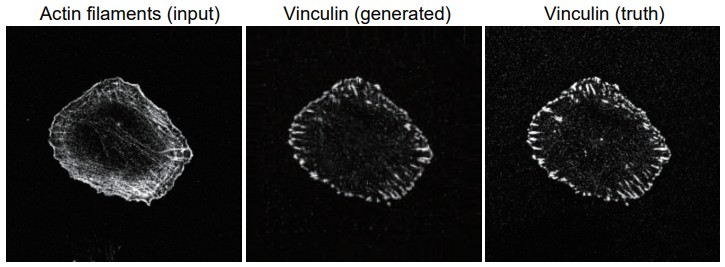
A example of a generated image of focal adhesion protein (vinculin) (center), that anchors actin filaments, from an image of actin filaments (left). The true vinculin image is also shown (right).
###
Resource
Title: Translation of cellular protein localization using convolutional networks
Authors: Kei Shigene, Yuta Hiasa, Yoshito Otake, Mazen Soufi, Suphamon Janewanthanakul, Tamako Nishimura, Yoshinobu Sato & Shiro Suetsugu
Journal: Frontiers in Cell and Developmental Biology
DOI: 10.3389/fcell.2021.635231
Information about project leader Suetsugu's lab can be found at the following website:
https://bsw3.naist.jp/eng/courses/courses210.html
( August 10, 2021 )
