Research outcomes
Genome-Maintenance Machine Captured!
High-speed atomic force microscopy (AFM) and yeast genetics unveil how a key protein complex in genome maintenance behaves dynamically
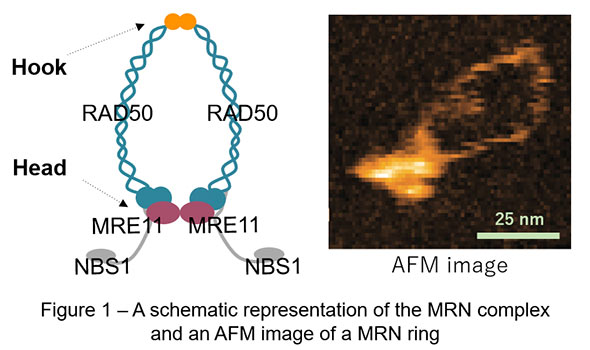
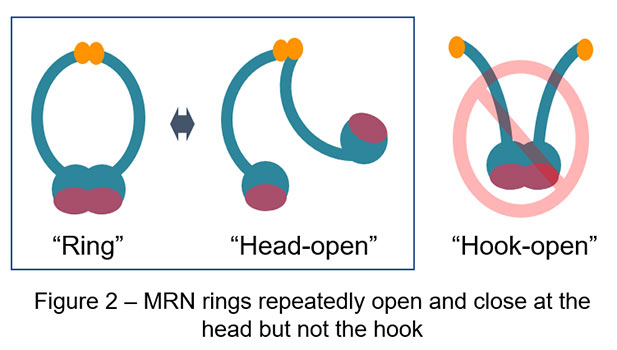
The MRE11/RAD50/NBS1 (MRN) complex is one of the key factors in genome maintenance and exists ubiquitously among species such as yeast and human. This complex contains two molecules of MRE11, RAD50, and NBS1, of which two RAD50 bind each other via both DNA-binding globular heads and hooks, exhibiting characteristic ring-like shapes (Figure 1). Previously, studies observing human MRN in atomic force microscopy (AFM) reported that human MRN rings frequently opened at the hooks, which led to a long-standing model that “hook-open” MRN holds broken DNA ends at DNA double-strand break sites by simultaneously binding DNA via the heads and forming higher order structures with adjacent MRN via the open hooks (Figure 2 – right panel). AFM used in these previous studies, however, was slow in image acquisition (approx. 30 sec / image) and hence practically incapable of observing dynamic motion of proteins in solution.
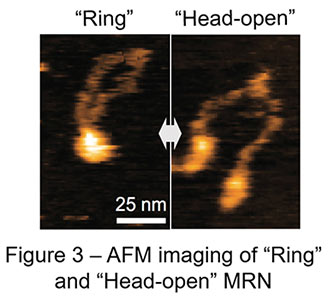
Recent advances have made AFM significantly faster in image acquisition (approx. 0.1 sec / image), which opens up the door for us to observe real motion of proteins in solution. In a collaboration study led by Assistant Professor Hisashi Tatebe at NAIST, Professor Takayuki Uchihashi at Nagoya University, and Associate Professor Asako Furukohri at Osaka University, this cutting-edge technology high-speed AFM is utilized to revisit dynamic motion of the MRN complex. Surprisingly, high-speed AFM in this study has captured totally different dynamic behavior of the MRN complex from the one previously reported and proposed: MRN rings repeatedly opens and closes at the heads instead of the hooks; MRN rings always stay closed at the hooks (Figure 2, 3). Genetic analysis in yeast has also found no harm at all in genome maintenance when the SMC hinge, another stable dimerization unit, replaces the Rad50 hook. Thus, rather than dynamically rearranging its binding, constitutive dimerization is the primary function of the RAD50 hook. This conclusion obtained by the combination of high-speed AFM and yeast genetics radically updates the framework how the MRN complex works in genome maintenance, bringing us solid ground in future development of cancer therapy as well as genome editing.
These latest results have been published in the journal of “Nature Communications” on January 17, 2020.
【Resource】
Tatebe, H., Lim, C.T., Konno, H., Shiozaki, K., Shinohara, A., Uchihashi, T., and Furukohri, A. (2020). Rad50 zinc hook functions as a constitutive dimerization module interchangeable with SMC hinge. Nat. Commun. 11, 370. doi: 10.1038/s41467-019-14025-0
URL:https://doi.org/10.1038/s41467-019-14025-0
【Cell Signaling Lab】
https://bsw3.naist.jp/eng/courses/courses304.html
Lab website: https://bsw3.naist.jp/shiozaki/
( February 12, 2020 )
