Research outcomes
Structural and functional analysis of the yeast N -acetyltransferaseMpr1 involved in oxidative stress tolerance via proline metabolism
The yeast Saccharomyces cerevisiae is an important microorganism not only in basic science as a model for higher eukaryotes like human, but also in fermentation industry for the production of breads, alcoholic beverages and bioethanol. Professor Hiroshi Takagi and his group members in the Laboratory of Applied Stress Microbiology have revealed that a novel N-acetyltransferase (NAT) Mpr1 mediates the L-proline and L-arginine metabolism by acetylating L-Δ1-pyrroline-5-carboxylate (P5C), leading to the L-arginine-dependent production of nitric oxide, which confers oxidative stress tolerance. Mpr1 has a unique primary structure and acetylates cyclic secondary amines as a substrate although known NATs acetylate primary amines, suggesting that Mpr1 has a novel 3D structure and catalytic mechanism.
In collaboration with Professor Hibi (Fukui Prefectural University) and Professor Hakoshima in the Laboratory of Structural Biology, Professor Takagi and his Graduate Student Ryo Nasuno presented the X-ray crystal structure of Mpr1 and its complex with the substrate cis-4-hydroxy-L-proline. They also demonstrated that Mpr1 recognizes its substrate by Asn135 and avoids steric interference due to the substrate binding by removing β-bulge structure, which is conserved among known NATs. Kinetic analyses using mutant enzymes revealed that Asn178 contributes to catalysis. Clarification of 3D structure and catalytic mechanism of Mpr1 provides new insights to understand the general structure-function relationship in NATs, which is involved in many important biological events. The structure-based design of improved mutant Mpr1 leads to the construction of industrial yeast with higher stress tolerance. Furthermore, the rational design of the specific inhibitor of Mpr1 may be also useful for the development of new antifungal drugs.
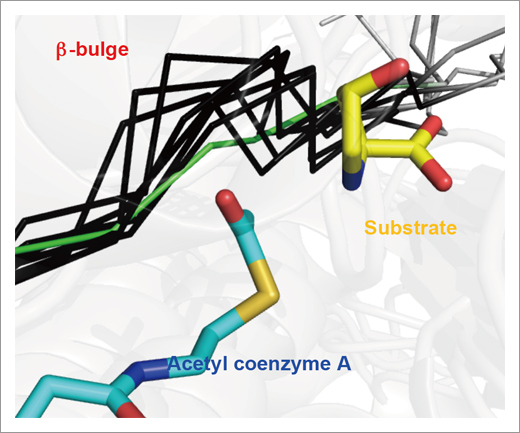
Fig. 1. The absence of b-bulge in Mpr1.
Mpr1 is shown by green and other typical NATs are shown by black. Mpr1 avoids the steric interference due to substrate binding by removing conserved b-bulge structure.
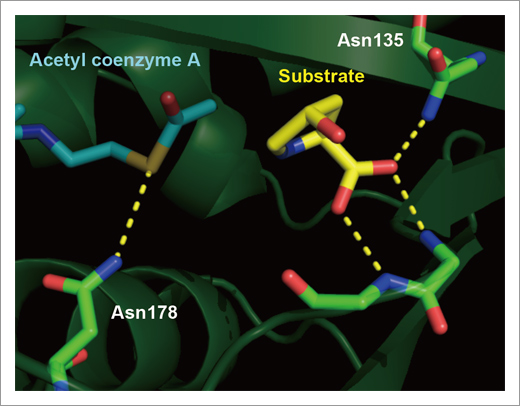
Fig. 2. The structure of active site on Mpr1.
Side-chain amide of Asn135 and backbone amide of Asn172 and Leu173 recognize a carboxyl group of the substrate (yellow), indicating that these residues are important for substrate recognition. And side-chain amide of Asn178 interacts with sulfur atom of acetyl-coenzyme A, suggesting that Asn178 catalyzes the enzymatic reaction of Mpr1.
[Press Release] July 2, 2013
( July 05, 2013 )
