PROJECT2
Development of Root Cap as a Root-Rhizosphere Interface
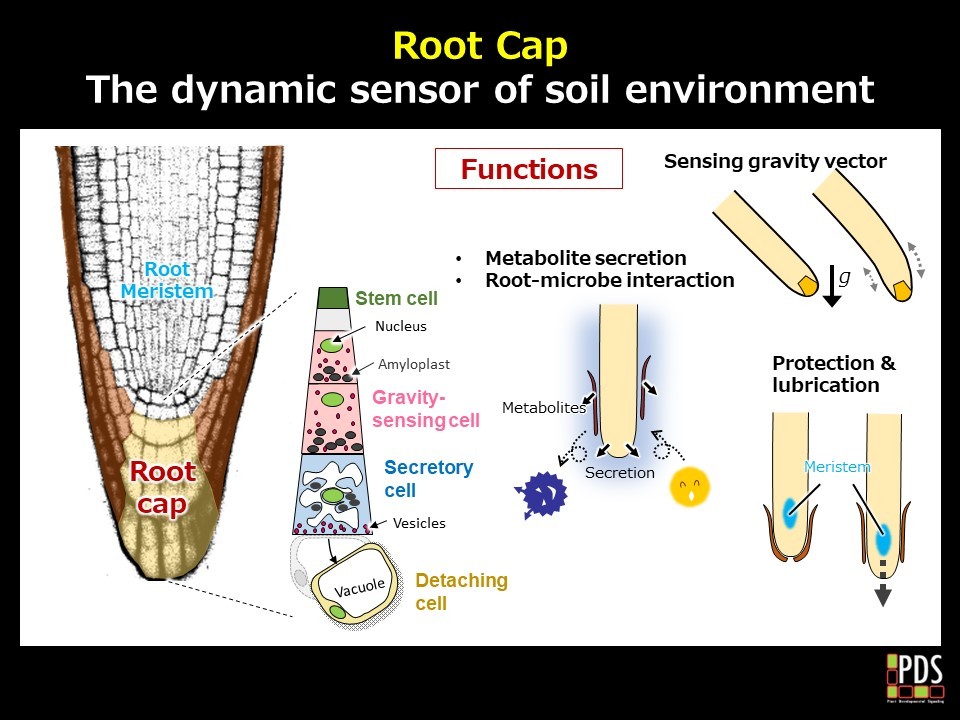
Taking the SMB/BRN transcription factor, the master regulators of root cap differentiation, as a starting point, we identified 60 genes that are likely to be involved in root cap formation and function. These genes encode enzymes acting on the cell wall, lipid synthases, and secretion-related factors. Our initial investigation centered around the RCPG gene, which is predicted to encode an enzyme that degrades pectin, one of the major components of the cell wall. Through live imaging, we found that RCPG protein rapidly increased in the outermost root cap cells, followed by the detachment of these cells. Mutants lacking RCPG function displayed incomplete detachment of the cell layer, causing the cells to remain at the root tip. This suggests that the cyclic activation of RCPG in the outermost layer of the root cap autonomously and periodically functions to release root cap cells.
Within the columella, which sits in the center of the root cap, the inner cells function as gravity-sensing cells, while the outermost cells function as secretory cells. Despite being identical columella cells, their intracellular structures are distinct. Our research indicates that autophagy is crucial in this transition process. While autophagy is a conserved intracellular self-digestion mechanism in eukaryotes, its role in plant tissue formation is largely unknown. Our study is unique not just for demonstrating the function of autophagy in plant development, but also for presenting a clear visualization of the process of cell differentiation at the root tip.