Project
Functional genomics by omics-based translational analysis
Plant secondary metabolism (also called “specialized metabolism”) produces compounds having several bioactivities such as resistance factors against various environmental stresses in plants, as well as health benefits for humans. Secondary metabolites are widely diversified in their chemical structures in nature (Fig. 1), since plants have adapted to environmental niches during long evolutionary periods using varied strategies such as gene duplication and convergent evolution of some key genes which contribute to chemical diversity. Our laboratory focuses on model plants, crop species and medicinal plants for i) the analysis of the natural diversity of secondary metabolites, and ii) the functional genomics approach by translational analysis of omics studies (genomics, transcriptomics and mass spectrometry-based metabolomics). The specific goal is the identification of key factors of natural chemical diversity and regulatory roles in plant secondary metabolism, which enable metabolic engineering of beneficial compounds.
Currently, after completion of full-genome sequencing of huge array of plant species, the complete biosynthetic framework needs to be applied for integrative approaches with other omics data such as genomics and transcriptomics. However, genome information is not sufficient to compute the size and framework of a species’ metabolism. Therefore, we screen qualitative differences of metabolite levels between tissues, stress treatments and wild accessions for “elucidation of biosynthetic framework” based on gene expression levels predicted from metabolite data (Fig. 2). On the other hand, recent technical developments allowing affordable whole genome sequencing, omics studies and availability of several resources such as knockout mutant library, quantitative trait locus (QTL) lines and wild accessions, have resulted in a dramatic increase in the number of approaches such as metabolic phenotype screening, network/modeling analysis, QTL and genome-wide association studies(GWAS). We therefore perform metabolomic analysis to screen qualitative differences of metabolite levels between different species, tissues and natural mutants for refinement of recent model of biosynthetic framework. Genomic and transcriptomic data as well as metabolomic data derived from genome-wide resources, will be used for translational analysis. We focus on the discovery of key genes involved in the creation of chemical diversity, and production of beneficial compounds.
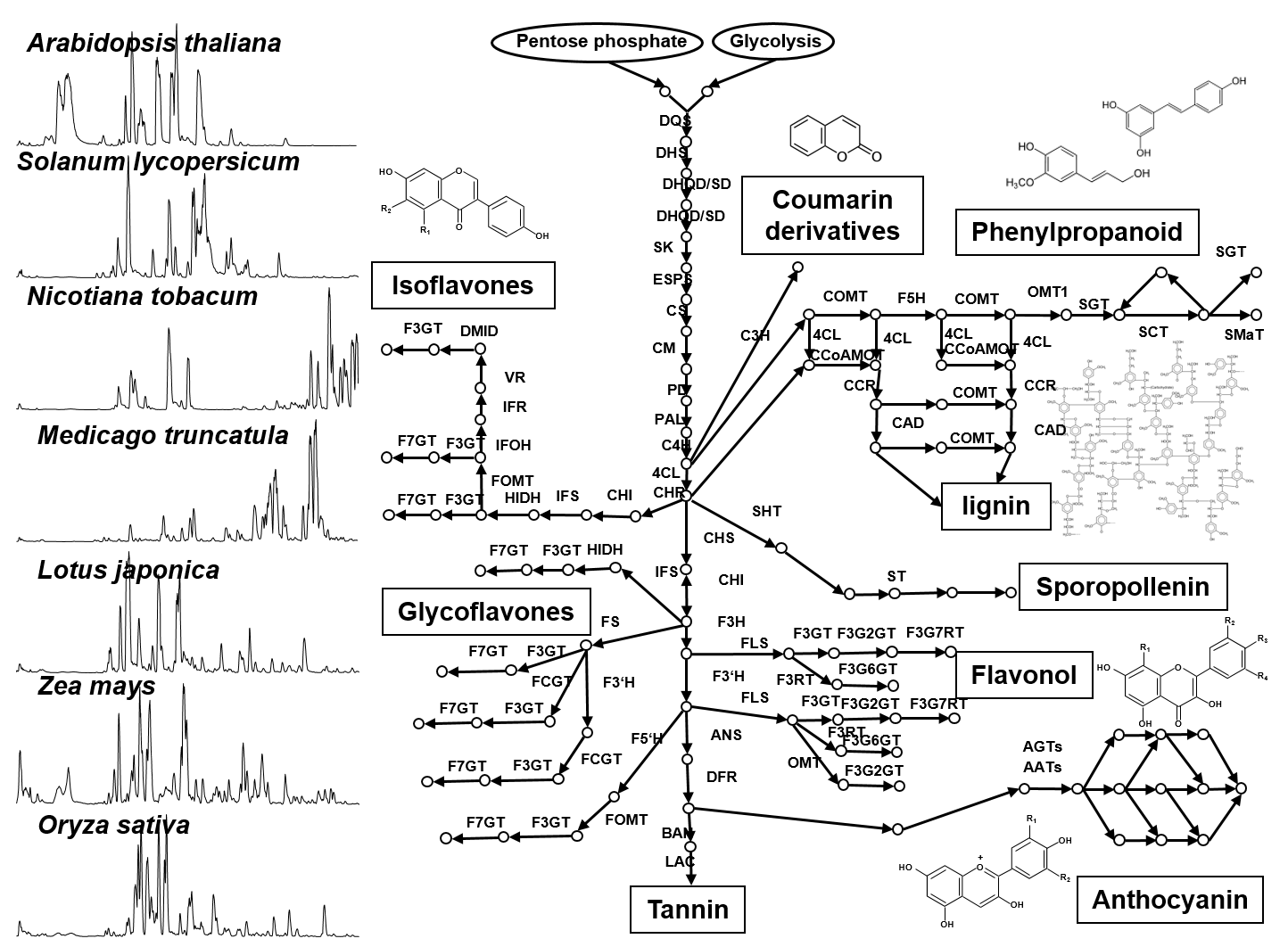
Fig.1 Metabolic network of plant polyphenolic biosynthesis and their chemical diversity between plant species.
On the other hand, recent technical developments allowing affordable whole genome sequencing, omics studies and availability of several resources such as knockout mutant library, quantitative trait locus (QTL) lines and wild accessions, have resulted in a dramatic increase in the number of approaches such as metabolic phenotype screening, network/modeling analysis, QTL and genome-wide association studies (GWAS)(Fig. 2). We employed these approaches supported by metabolomic studies to refine biosynthetic framework of plant secondary metabolism including natural variance, tissue and species specificity, in order to discover key genes involved in the creation of chemical diversity.
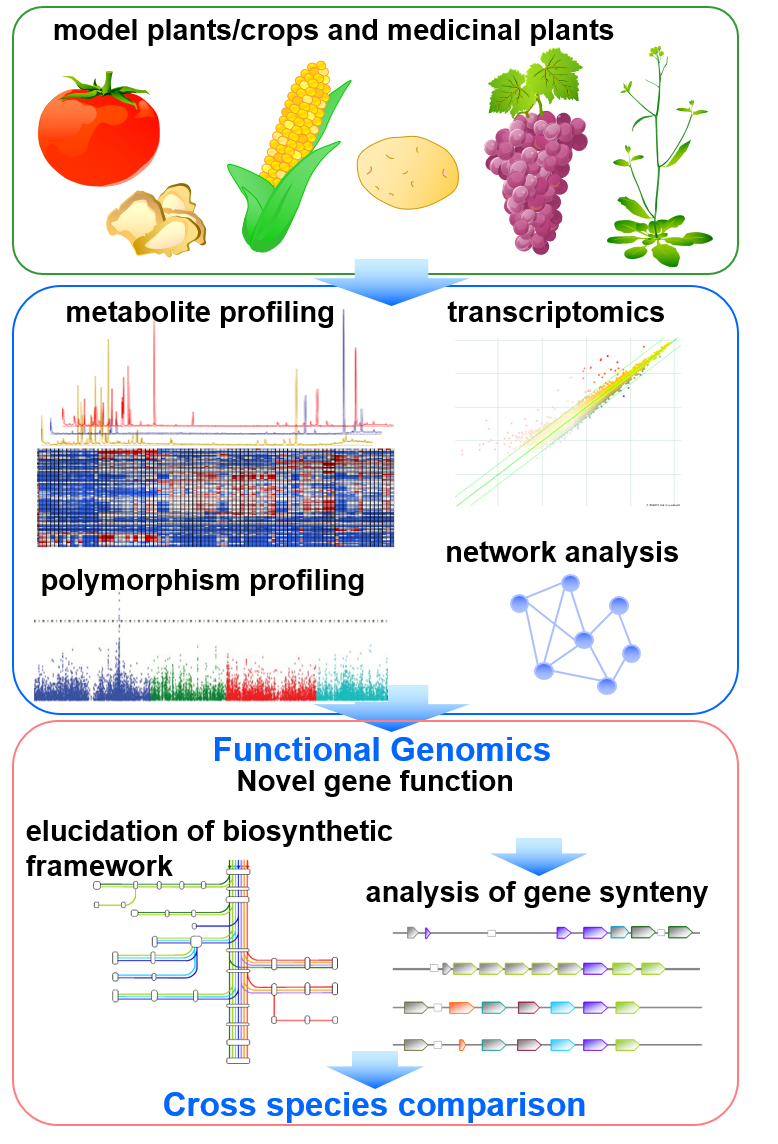
Fig.2 Omics-based translational analysis using model plants and crops.
Cross species comparison on the neo-functionalized genomic region
Recent technical developments allowing reasonable whole genome sequencing as well as a better inventory of species-by-species chemical diversity have greatly advanced our understanding as to how these pathways vary across species. The range of genetics based strategies for characterization of key genes described above provide several genes and genomic regions involved in neo-functionalization of plant secondary metabolism. Neo-functionalization” which occurs a totally new function after a gene duplication event, is a key factor of functional gen divergence. We therefore focus on the species specific duplicated genes in these key genome synteny regions in order to discover new functional genes in plant secondary metabolism.
Regulation of metabolic network during nutritional stress
Plant growth and development highly depend on the availability of nutrients. Nutrient deficiencies in soils cause severe reduction in growth with low yield and quality of crop, which is one of serious problems in agriculture. To cope with the nutrient deficiencies, plants have evolved several adaptation strategies. Understanding of these strategies at the molecular level is required for effective breeding of plants having high nutrient use efficiency or tolerance to nutritional stress. We investigate the metabolic and gene expression changes of plants grown under nutrient deprivation stresses. The purposes of this study are: i) making an index of time-dependent metabolic changes, ii) evaluating the robustness of metabolic network, iii) finding species-conserved metabolic makers, for effective breeding of plants having high nutrient use efficiency or tolerance to nutritional stress.

