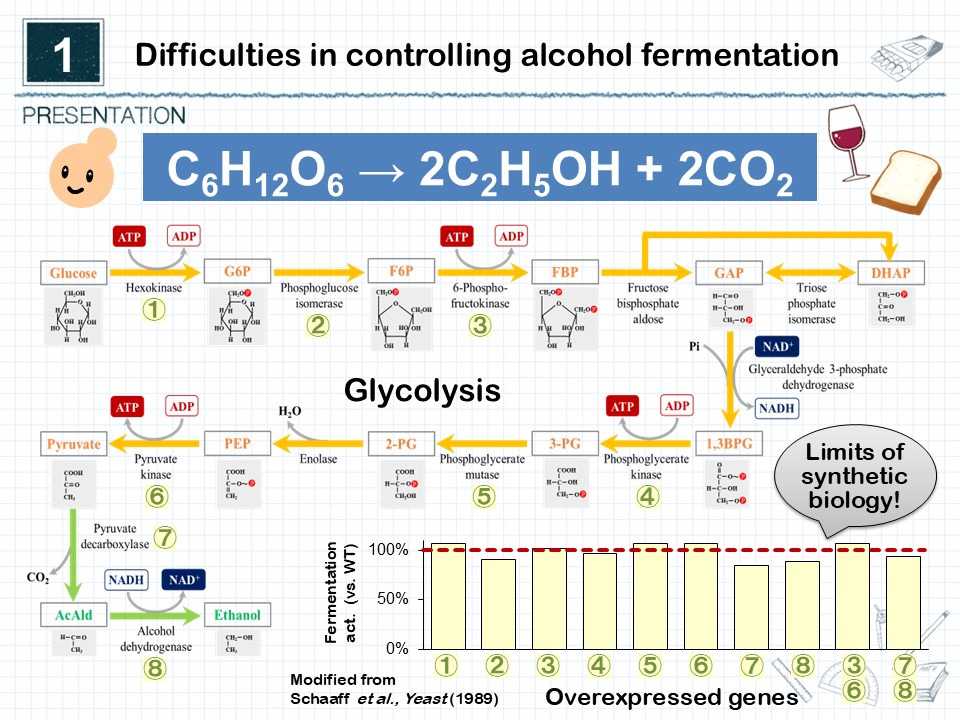
Yeast alcoholic fermentation is one of the most familiar microbial metabolic functions to humankind, having been utilized since prehistoric times for the production of alcoholic beverages, fermented foods, and bioethanol. Decades of research have elucidated all the enzymes that catalyze each metabolic reaction, as well as the genes encoding them. In modern times, synthetic biology has advanced to the point where bioengineering technologies for enhancing the production of valuable compounds are becoming universal methodologies. However, modifying yeast alcoholic fermentation remains surprisingly difficult, and no clear breakthrough has been achieved (Fig. 1). In the fermentation and brewing industries, traditional techniques that control yeast viability solely through temperature and nutrient management are still widely used, and optimizing the alcoholic fermentation capacity per cell remains a major challenge.
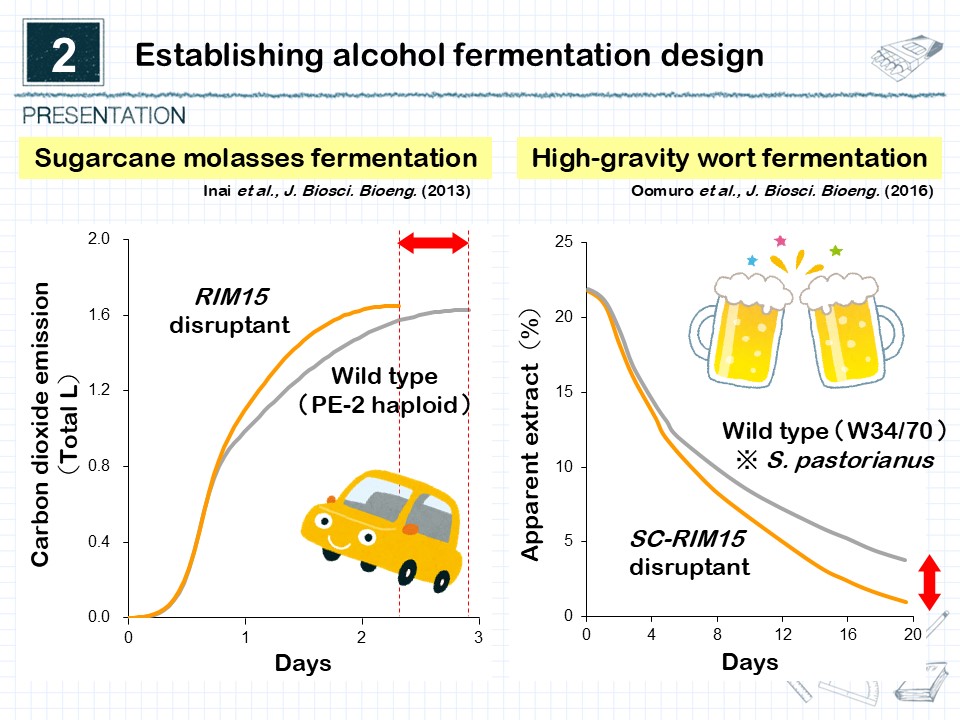
We focused on the high alcoholic fermentation capacity of sake yeast, a valuable microbial resource unique to Japan. Omics analyses have revealed specific mutations found only in sake yeast. Introducing these mutations into laboratory yeast strains with low alcoholic fermentation capacity enabled them to achieve fermentation performance comparable to sake yeast. By applying this technology, it becomes possible to precisely modify the alcoholic fermentation ability of various yeasts used in the fermentation and brewing industries (Fig. 2). Indeed, we have successfully enhanced the alcoholic fermentation capacity of bioethanol yeast and brewer’s yeast. Conversely, by suppressing the alcoholic fermentation of sake yeast, we have also developed yeast strains suitable for producing low-alcohol sake. Moving forward, we aim to further identify key genes that regulate alcoholic fermentation and establish a unique “alcoholic fermentation design technology” through their modification.
【Related Papers】
- D. Watanabe*, M. Kawashima, N. Yoshioka, Y. Sugimoto, and H. Takagi; Rational design of alcoholic fermentation targeting extracellular carbon. NPJ Sci. Food 7(1): 37 (2023)
- D. Watanabe*, T. Kajihara, Y. Sugimoto, K. Takagi, M. Mizuno, Y. Zhou, J. Chen, K. Takeda, H. Tatebe, K. Shiozaki, N. Nakazawa, S. Izawa, T. Akao, H. Shimoi, T. Maeda, and H. Takagi; Nutrient Signaling via the TORC1-Greatwall-PP2AB55δ pathway is responsible for the high initial rates of alcoholic fermentation in sake yeast strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 85: e02083-18 (2019)
- D. Watanabe, Y. Zhou, A. Hirata, Y. Sugimoto, K. Takagi, T. Akao, Y. Ohya, H. Takagi, and H. Shimoi*; Inhibitory role of Greatwall-like protein kinase Rim15p in alcoholic fermentation via upregulating the UDP-glucose synthesis pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 82: 340-351 (2016)
- D. Watanabe, Y. Araki, Y. Zhou, N. Maeya, T. Akao, and H. Shimoi*; A loss-of-function mutation in the PAS kinase Rim15p is related to defective quiescence entry and high fermentation rates of Saccharomyces cerevisiae sake yeast strains. Appl. Environ. Microbiol. 78: 4008-4016 (2013)
- D. Watanabe, H. Wu, C. Noguchi, Y. Zhou, T. Akao, and H. Shimoi*; Enhancement of the initial rate of ethanol fermentation due to dysfunction of yeast stress response components Msn2p and/or Msn4p. Appl. Environ. Microbiol. 77: 934-941 (2011)
