Pfof. K. KATO
Isolation and improvement of elements involved in high expression of transgene
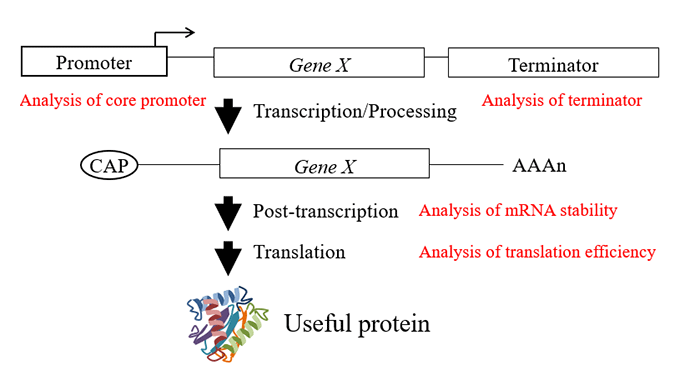
Gene expression is controlled by several processes such as transcription, post-transcription, and translation. In order to efficiently express the introduced useful genes in plants, it is necessary to optimize each process. Therefore, we are analyzing sequences related to the efficiency of control processes such as core promoter, terminator, splicing site, 5'UTR and 3'UTR. Through these analyzes, we are isolating and improving the sequence elements involved in high expression. We also aim to provide the results to various companies to produce useful proteins such as vaccine proteins and growth hormones in plants.
Assist. Prof. T. TAMURA
--- under construction ---

--- underconstruction ---
Assist. Prof. T. WAKABAYASHI
Phenotypic and genetic polymorphisms in plants
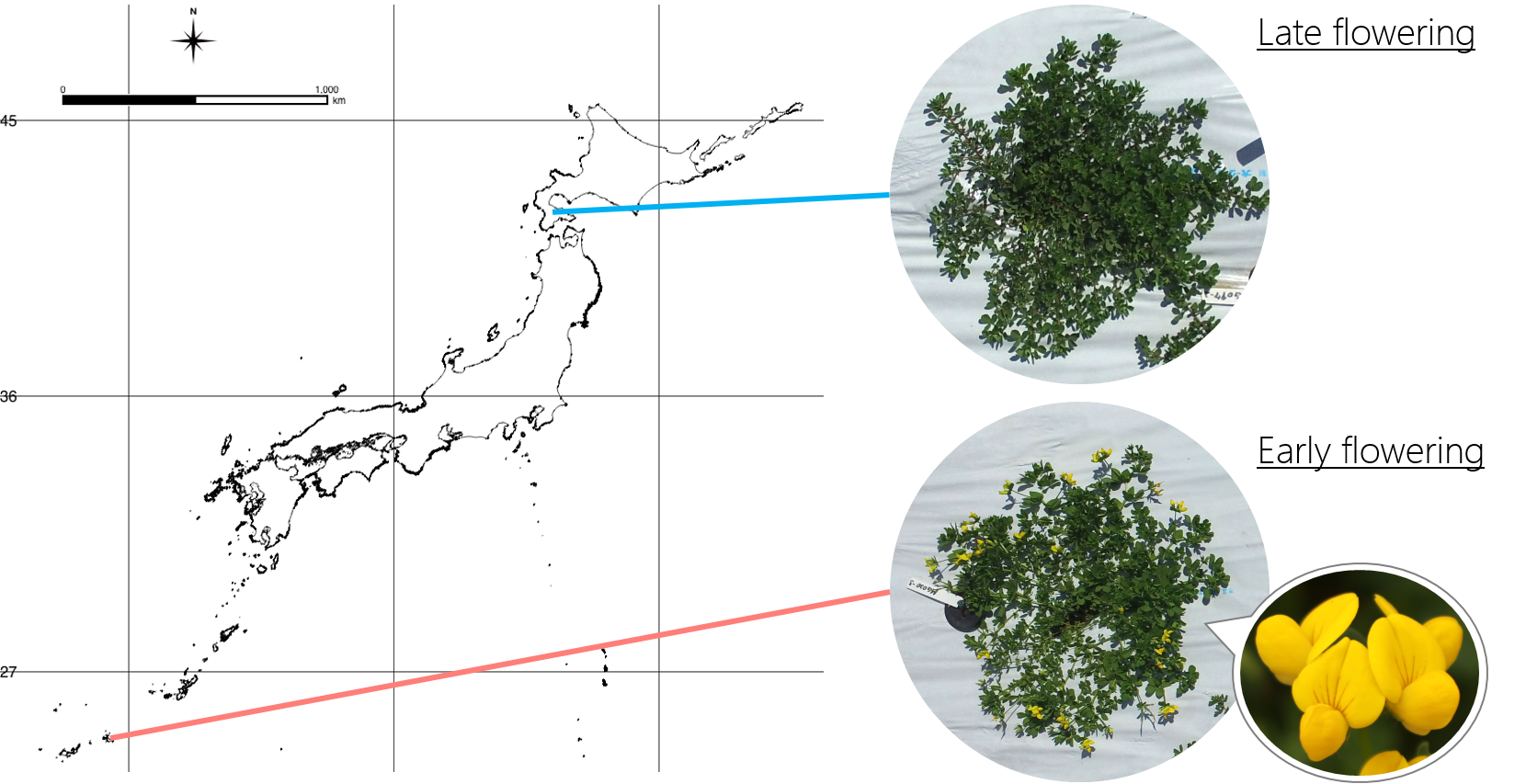
Nucleic and phenotypic polymorphisms are maintained even in a species. Especially for plant species, which do not move from the place to another, natural selections strongly affect to its genotype and phenotype in the process of local adaptation. Therefore, these polymorphisms are important to persistence and migration of the species. In addition to wild plants, intraspecific polymorphisms are also important in agricultural varieties. There are various polymorphisms in agricultural varieties and these polymorphisms could be keys of breeding for their specific characteristics including tastes and disease resistances.
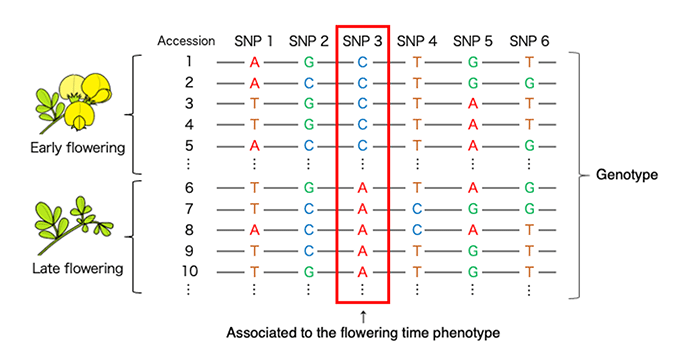
Putting both of phenotypic polymorphisms and genome-wide nucleotide variation, which can be gathered by next-generation sequencing technology, into association analyses, related gene regions can be detect. We are focusing on traits related to the plant reproductive success, for example, flowering time and annual/perennial, because these traits are directly related to biodiversity and breeding.
Assist. Prof. T. KATO
Elucidation of the environmental adaptation mechanism of plants that survive harsh environments

We focus on the ability of plants to cleverly withstand harsh environments (particularly water stress)
and the mechanisms by which they flexibly adapt to environmental changes. We will conduct research mainly using the liverwort,
Marchantia polymorpha, which grows in a wide range of environments, mainly in Temperate humid region.
Currently, we are using strains with different environmental adaptability to search for responsible genes through genetics and genome analysis,
and are also focusing on the mechanism by which plants that have stopped development due to dryness can resume development when they receive water again.
By combining high-resolution phenotypic analysis and gene expression analysis, we hope to understand plant cells that are resilient to environmental changes.
Bryophytes are the first to branch out at the base of land plants, and it is hoped that mechanism of drought tolerance
that is common to land plants or unique to bryophytes, will be discovered.