
RESEARCH
PRR-mediated signal transduction
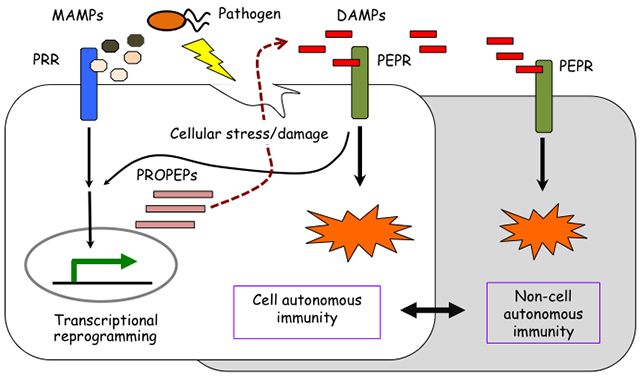
The surfaces of plant cells harbor an array of PRRs that monitor the extracellular spaces for microbial MAMPs and self-derived DAMPs. The two major domain structures are overrepresented in the ligand-binding ecto-domains of PRRs and co-receptors: the leucine-rich repeat (LRR) and lysine-motif (LysM) domains. In Arabidopsis, the LRR receptor kinases FLS2 (flagellin sensitive-2) and EFR (EF-Tu receptor) act as the receptors for bacterial flagellin (flg22 epitope) and EF-Tu (elongation factor thermo unstable, elf18 epitope), respectively. The LysM receptor kinase CERK1 (chitin elicitor receptor kinase 1) functions as the receptor for chitin, a major component of fungal cell walls, and is also essential for perception of bacterial peptidoglycans. The LRR receptor kinases PEPR1 and PEPR2 are DAMP receptors that recognize endogenous PROPEP elicitor peptides (Pep epitopes). Ligand binding to PRRs almost instantly induces the formation of a receptor complex(es) with co-receptors and signaling adaptors, which provides a platform for triggering intracellular signaling. Sensing of different MAMPs/DAMPs typically triggers a series of cellular outputs: changes of ion fluxes across the plasma membranes, a reactive oxygen species (ROS) burst, and MAPK activation are typically detectable within minutes, with ethylene production, transcriptional reprogramming, cell wall remodeling (e.g., callose deposition) and metabolic changes following within several hours to days. Taking advantage of the ease of tracking these proxies in Arabidopsis, we aim to elucidate the mechanisms by which a single receptor governs and coordinates separate signaling outputs, a common theme in biology.
Until recently, PRRs were thought to elicit a common set of defense responses, irrespective of the type of ligands. However, we found that individual PRR signaling systems have their own advantages and rate-limiting steps, and complement one another to strengthen the overall immune response. We seek to understand the significance and mechanisms of signaling crosstalk between separate PRR-mediated pathways in plant immunity.
Although individual cells are assumed to act as defensive units against pathogen infection, cell–cell crosstalk also plays important roles in the control of overall plant immunity. For example, a danger signal generated from pathogen-challenged cells can be sensed not only in the infected cells but also in the surrounding cells. Such cell–cell communication is likely to provide a basis for both propagation and containment of defense signaling, although the underlying process remains poorly understood. We aim to reveal the steps of defense signaling that are controlled in a cell-autonomous manner (i.e., by individual cells) or a non-cell-autonomous manner (i.e., by intercellular communication).

Utilization of DAMP signals and receptors to boost plant defense
To enable sustainable crop production, we must deepen our understanding of the plant immune system and apply this knowledge to disease control and promotion of plant growth with the aid of beneficial microbes. Conventional techniques for plant disease management, which generally rely on R genes or antimicrobial substances, have significant disadvantages, such as a narrow spectrum of target pathogens and short-lived resistance due to the rapid evolution of pathogenic species. We seek to overcome those problems by taking advantage of the PRR-mediated immune regulatory network, with a particular focus on DAMP signals and receptors. We expect that a trade-off with growth can be minimized by exploitation of DAMP signaling systems, whose activation is essentially specific to pathogens and leads to host resistance against a broad spectrum of pathogens.
Signal integration between immune and environmental responses in plants

In nature, plants often need to mount immune responses against pathogen attacks while dealing with a combination of different abiotic stress cues from the environment. However, little is known about the strategies and mechanisms by which plants accomplish this coordination during simultaneous exposure to different stress cues. Plants often adopt a prioritized strategy, dealing with a particular stress or stresses while postponing or suppressing the response to others. The phenomenon of crosstalk is widespread in higher plants, in which MAMP signaling activation overrides sucrose/UVB-stress induced flavonoid accumulation. (We exploited this feature as a readout in our previously reported screens for MAMP-insensitive psl mutants in Arabidopsis.) Flavonoids have a diverse array of physiological functions, not only acting as antioxidants and/or sunscreen pigments to protect plants from oxidative and UV light damage, but also as modulators of auxin transport. Our work has revealed antagonism between flavonoids and salicylic acid (SA)-mediated immunity, highlighting the importance of fine-tuning flavonoid metabolism as a critical step in adaptation to different biotic and abiotic stresses. Thus, this system is suitable for studies on signal integration between biotic and abiotic stress responses.Building on these findings and considerations, we initiated a study of crosstalk between immune responses and environmental factors, such as light, water (humidity), temperature, and nutrition. In this manner, we explore the mechanisms by which plants optimize the balance between growth, immune, and environmental responses.
Histone-mediated regulation of immunity-related gene expression in plants
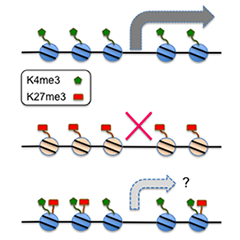
It is assumed that individual plant cells are equipped with both PTI and ETI. PTI is induced when PRRs recognize microbial components or endogenous elicitors (MAMPs and DAMPs), whereas ETI is induced when R proteins, of which the majority are NLR (nucleotide-binding domain LRR) receptors, recognize pathogenic effectors (virulence factors). Local activation of PTI or ETI at primary pathogen-challenged sites in turn heightens the state of cellular immunity in distant, non-challenged sites, a phenomenon termed systemic acquired resistance (SAR). SAR is typically followed by a “primed” state, in which target genes are poised for enhanced, sensitized, and/or faster activation upon subsequent pathogen challenge. Our unpublished data and recent work by others imply the engagement of histone modifications, which are typically associated with transcriptional memory, in priming of defense-related genes. Defense priming introduces a condition of elevated readiness that enables effective resistance when plants are attacked again, without imposing the huge costs required for full defense activation. A primed state may last across multiple seasons or generations. Successful exploitation of defense priming is expected to solve the trade-off problem with plant growth in breeding of biotic/abiotic stress-tolerant plants.Our pilot studies indicate that certain types of histone modifications at primed target genes and histone modifiers play important roles in systemic defense priming, in particular after primary ETI activation. We are currently exploring the mechanisms by which plants establish and maintain a memory of defense-related transcriptional reprogramming as a defined chromatin state(s). This line of research is expected to unveil the mechanisms by which plants simultaneously activate and deactivate a wide array of defense-related genes.
Infection strategies of endophytic and pathogenic microorganisms, and the plant immune response
Countless endophytic or symbiotic microorganisms inhabit plant tissue without discernible disease symptoms. Some endophytes assist the host with nutrient uptake and pathogen resistance. However, we still have a great deal to learn about roles of these organisms in plant eco-physiology and pathology. For fungal endophytes, arbuscular mycorrhizal fungi, which are present in an estimated 80% of land plants, are the traditional model for research. However, the lack of culture methods and genetic manipulation procedures for mycorrhizal fungi, and for fungal colonization in Arabidopsis, represent major bottlenecks in performing genetic studies of mycorrhizal symbiosis.
To overcome these hurdles, we propose to employ the endophytic fungus Colletotrichum tofieldiae in systematic genetic studies of plant-endophyte interactions. This root endophyte species was first isolated from wild populations of Arabidopsis in Spain. C. tofieldiae is prevalent and promotes plant growth in phosphorus- and nitrogen-deficient conditions. Whole-genome sequencing revealed that C. tofieldiae is closely related to the canola pathogen C. higginsianum and the corn pathogen C. graminicola, indicating a close phylogenetic relationship between many endophytes and pathogens. From the green field of NAIST, Japan, we have also identified many more Colletotrichum isolates/species with different characteristics. We will utilize a collection of endophytic and pathogenic members of the genus Colletotrichum to uncover the similarities and differences in their infection strategies. We will also elucidate the mechanisms by which plants discriminate pathogens and non-pathogens by comparing the host immune responses to these organisms (e.g., by analyzing DAMP-mediated responses).