
植物代謝制御研究室
Laboratory of Plant Metabolic Regulation
Research
当出村研究室では、持続可能な社会の構築に向けて、エネルギー生産、環境再生、食糧増産に役立つ植物の創出と活用に関する研究と教育を行っています。 モデル植物や実用植物のオミクス情報をもとに分子生物学的な解析手法とバイオテクノロジーを用いて、植物細胞の分化制御機構の解析、植物の機能と代謝の調節機構の解析、有用トランスジェニック植物・樹木の作出を進めています。
Lab Tour
出村教授による植物代謝制御研究室の実験機器や研究材料の紹介です。
Bioscience Discovery Session
出村教授による研究内容の紹介です。
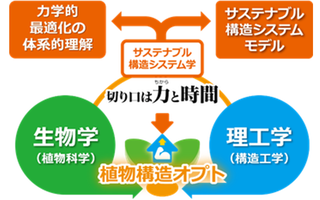
Research
当研究室の研究テーマの紹介
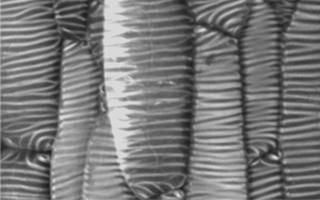
Publication
発表した論文や学会発表の紹介

Blog
研究室のブログ



