Elucidating the detailed mechanisms of motor protein SecDF in protein translocation across the membrane.
A proposed new mechanism for protein translocation across the membrane: Opening and closing mechanism of the cap of the protein-conducting channel
Furukawa A, Yoshikaie K, Mori T, Mori H, Morimoto VY, Sugano Y, Iwaki S, Minamino T, Sugita Y, Tanaka Y, *sukazaki T. tunnel formation inferred from the I-form structures of the proton-driven protein secretion motor SecDF cell rep. 19, 895-901 (2017).
Summary
The cell membrane separates the cell’s interior from the environment. Extracellular transport of proteins synthesized in the cytoplasm is an essential process for sustaining life. We conduct basic research on the molecular mechanisms of the protein transport.
In bacteria such as Escherichia coli, the membrane-embedded SecDF is involved in protein transport. Specifically, SecDF mediates the translocation of its substrates by using the H+ gradient across the membrane. However, the detailed molecular mechanisms of SecDF remains unclear. In particular, the sites that interact with substrate proteins passing through the membrane and the H+ pathway remained elusive.
Recently, we successfully determined the crystal structure of SecDF at high resolution (2.6 Å). Based on this structural information, we carried out biochemical experiments and computational molecular dynamics simulation. The results of these analyses identified SecDF’s a substrate-binding site and a H+ ion pathway. Moreover, we demonstrated that the each SecDF domain undergoes dynamic conformational changes, driven by the H+ ion gradient. Based on these findings, we proposed a novel molecular mechanism for SecDF-driven protein translocation across the membrane.
Our findings regarding this essential biological phenomenon will contribute to progress in basic life science. In addition, our findings will provide a theoretical basis for developing new antibiotics that target the bacteria-specific SecDF protein.
Study Description
(A) Background
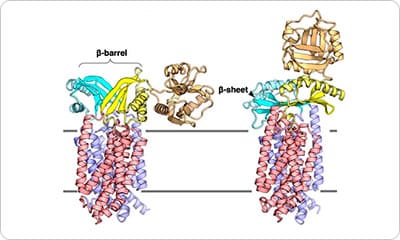
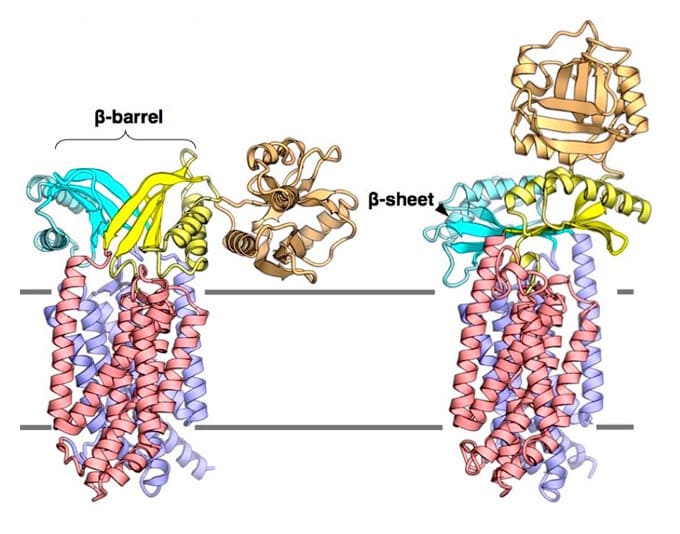
Cellular compartments are distinctly separated from their exterior environment by the membrane. Although the cell membrane does not allow free passage of molecules and ions, their bidirectional transport across the membrane is necessary for maintaining life. Proteins, which are synthesized by cytoplasmic ribosomes, are necessary components of living organisms. Approximately 30% of proteins are translocated across the cell membrane to perform various activities in the extracellular milieu. Protein translocation is mediated by the ubiquitous Sec translocon channel. Because the Sec translocon functions as a passive channel, accessory proteins must assist in protein secretion. In bacteria, the motor proteins SecA and SecDF drive the polypeptide translocation. SecDF is a bacteria-specific membrane protein consisting of transmembrane and periplasmic domains. It is widely believed that along with the influx of H+ through the transmembrane domain, SecDF undergoes repeated cycles of conformational transition from the F-form (capturing state) to the I-form (holding state) in order to undergo translocation of polypeptides released from the Sec translocon to the periplasm. However, previous studies reported only the full-length F-form of the SecDF structure, underscoring the need for high-resolution structure of SecDF in I-form .
(B) New Findings
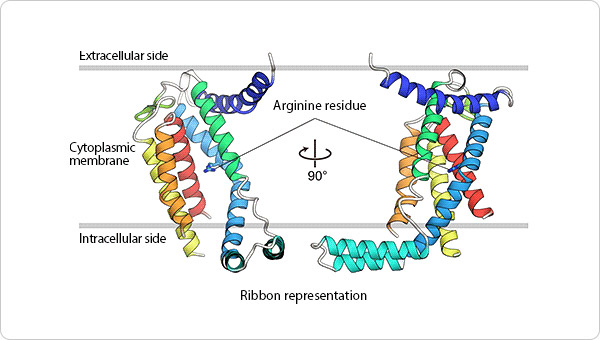
We successfully determined the crystal structures of I-form SecDF from Deinococcus radiodurans, at a resolution of 2.6 Å. The lipidic cubic phase (LCP) crystallization technique was applied to crystallize SecDF, and X-ray diffraction data sets were collected at beam line BL32XU at SPring-8, Hyogo, Japan.
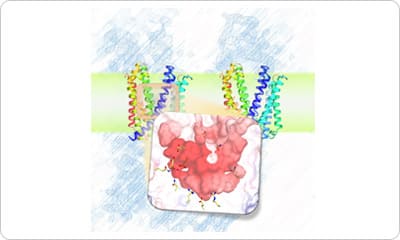
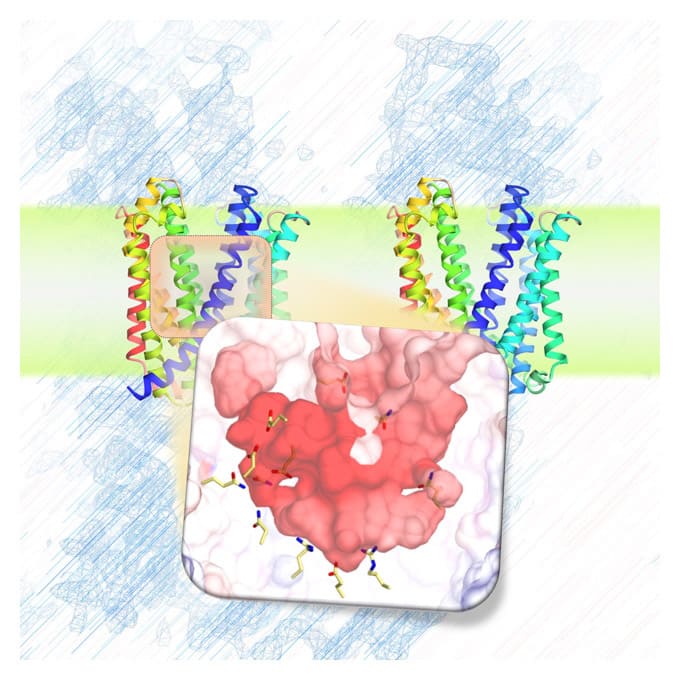
The I-form SecDF structure exhibits two noteworthy characteristics. First, the cavity of the P1 domain, the first periplasmic domain, was occupied by a small molecule, probably mimicking the binding of substrate protein. We conducted a biochemical study to identify the interaction site in SecDF with the protein substrate, and found that a substrate-specific interaction site is located in this cavity. These results strongly suggest that the cavity forms the substrate-binding site.
Second, the transmembrane region of SecDF possesses a tunnel architecture that spans across the membrane form the cytoplasm to periplasm. This tunnel seems to be the H+ ion pathway, because an aspartate residue essential for SecDF functions (i.e., proton transfer and preprotein translocation activation) was located at the center of the tunnel. We performed molecular dynamics simulations of SecDF; the results implied that the deprotonated and protonated states of the aspartate residue are closely related to the opening and closing of the tunnel, respectively, and that water molecules are aligned via hydrogen bonding along the inner surface of the tunnel ranging between its cytoplasmic and periplasmic ends. These results suggest that the tunnel allows for H+ ion entry into the cytoplasm from the periplasm. Moreover, we found that H+ ion influx and preprotein translocation are prohibited when the highly flexible P1 domain is rigidly fixed. This finding suggested an association between P1 domain flexibility and H+ ion conductance activity.
Based on these findings, we proposed a new working model for SecDF: i) the F-form conformation of SecDF captures the substrate protein in the P1 cavity; ii) SecDF undergoes the conformational transition from the F- to the I-form with the substrate. the conformational F-to-I transition is driven by energy derived from the H+ ion influx, facilitated by water molecules in the transmembrane domain tunnel; and iii) preprotein translocation is achieved by repeated cycles of the conformational F-to-I transition.
(C) Significance and Future Perspectives
In this study, we revealed the molecular mechanisms of SecDF motor. Elucidation of the protein translocation mechanisms, which are essential for life, will greatly contribute to basic research in related fields. Because SecDF is specific to and essential for bacterial survival, knowledge of SecDF mechanisms will provide a technical framework for the discovery of new antibacterial agents.


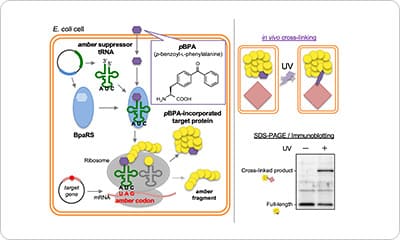
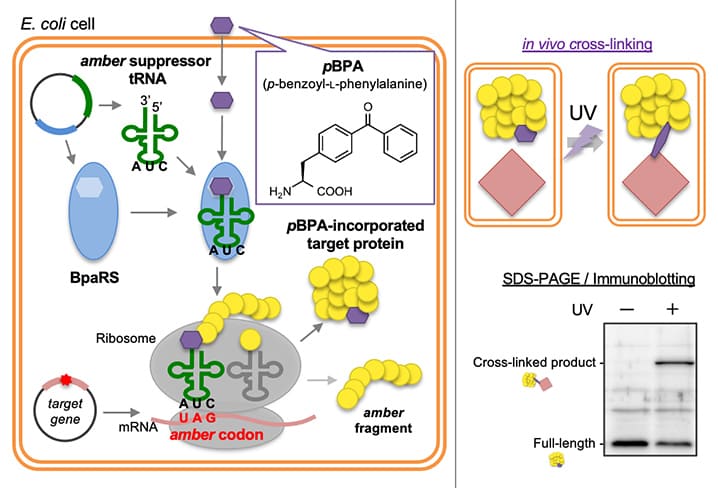




Highlights